1. Introduction
Histone deacetylases (HDACs) are a group of enzymes that play a crucial role in the regulation of gene expression. They are involved in the modification of histones, which are proteins that help package DNA into a compact structure called chromatin. The acetylation and deacetylation of histones are dynamic processes that influence the accessibility of DNA to transcriptional machinery, thereby affecting gene expression [
1,
2,
3]. HDACs or EC 3.5.1.98 comprise a category of enzymes responsible for eliminating acetyl groups (O=C-CH
3) from the ε-N-acetyl lysine amino acid found on both histone and non-histone proteins [
4]. Inhibitors of histone deacetylase represent a novel category of cytostatic agents designed to impede the proliferation of tumor cells [
5,
6,
7].
This brief theoretical study seeks to explore the potential role of Curcumin, a natural substance recognized for its anti-tumor properties [
8,
9], through an in silico approach employing Molecular Docking [
10]. The objective is to ascertain whether Curcumin demonstrates effective binding to Histone deacetylases, evaluating the binding affinity and identifying the specific types of bonds involved in the interaction.To accomplish this objective, we utilized the Mcule Database [
11], leveraging the Autodock Vina algorithm for an automated exploration of the role of Curcumin in binding interactions.
2. Material and Methods
Several Histone deacetylases are performed in this work:
- -
Histone deacetylase 8 ( PDB Code 3mz6) : Binding site center (ångström) x( 0,1808), y( 42,7338) z( 2,1861)
- -
Histone deacetylase 6 (PDB Code 3gv4):Binding site center(ångström) x( -7,3659), y( 4,9697) z( 0,9734)
- -
Histone deacetylase-like amidohydrolase ( PDB Code1zz1) :Binding site center(ångström) x( -28,5197), y( 16,2263) z( -16,5053)
- -
Histone deacetylase 4( PDB Code 2vqq) Binding site center (ångström) x( -17,5922), y( 7,3406 z( -11,4742)
- -
Histone deacetylase 2( PDB Code 3max) Binding site center (ångström) x( 48,9732), y( 15,6496z( -43,4309)
3. Results and Discussion
This study specifically delves into the potential interaction of Curcumin with Histone deacetylases using Molecular Docking via the Mcule Database. Molecular docking is a computational technique that simulates the interaction between molecules [
10], such as a ligand (e.g., Curcumin) and a protein (e.g., Histone deacetylase). Binding energy is a critical parameter in these studies, reflecting the strength and stability of the interaction between the ligand and the protein's binding site.
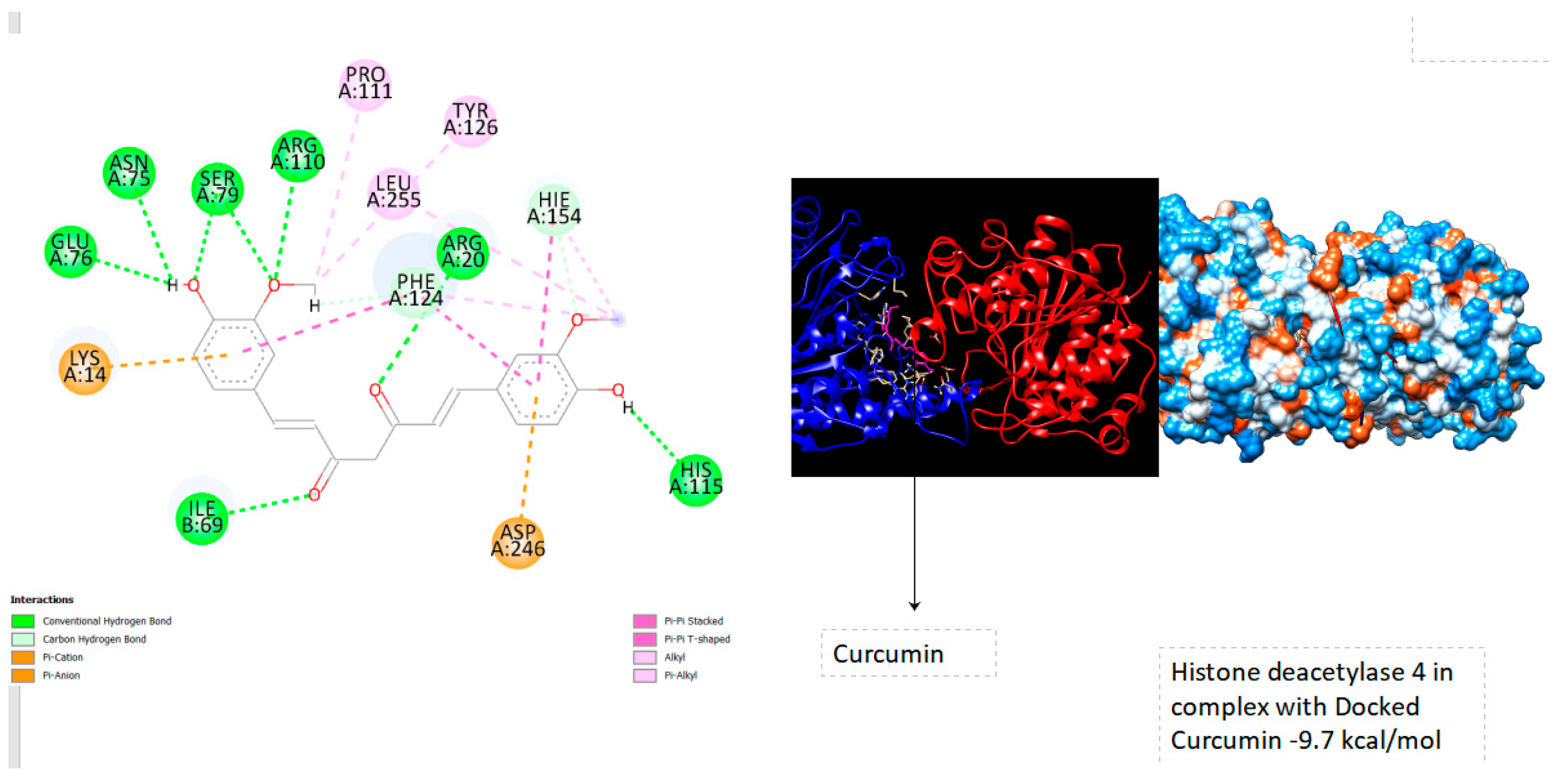
In general, a lower binding energy indicates a more favorable and stable binding interaction. Therefore, when comparing binding energies across different Histone deacetylase targets, researchers can identify which one has the strongest binding affinity for Curcumin. This information is valuable in understanding the potential effectiveness of Curcumin as a ligand for different protein targets and can guide further exploration of its therapeutic applications. Notably, the results reveal a notably elevated binding energy of -9.7 kcal/mol within the Ligand Binding Site of Histone deacetylase 4 (See below
Figure 1). In
Table 1 was shown the comparison binding energies of Histone deacetylases targets with Curcumin.
Histone Deacetylase 4 (HDAC4) is a gene that encodes a protein involved in regulating gene expression through the deacetylation of histones. The deacetylation of histones by HDAC4 is part of the larger process of epigenetic regulation, influencing how genes are turned on or off.
HDAC4, as a histone deacetylase, is involved in the regulation of gene expression through transcriptional processes. Transcription is the synthesis of RNA from a DNA template, a key step in gene expression.
Understanding the role of HDAC4 in these pathways and its association with specific disorders provides insights into the broader impact of epigenetic regulation on human health and development. Additionally, these findings contribute to the exploration of potential therapeutic targets for conditions linked to HDAC4 dysfunction [
12,
13,
14].
The binding energies (in kcal/mol) for Curcumin with different Histone deacetylases are as follows:
Histone deacetylase 8: -6.4
Histone deacetylase 6: -6.5
Histone deacetylase 2: -8.4
Histone deacetylase 4: -9.7
Histone deacetylase-like amidohydrolase: -5.8
The results indicate that Curcumin exhibits a higher affinity for Histone deacetylase 4 compared to other tested histone deacetylases. The more negative the binding energy, the stronger and more stable the interaction, suggesting that Histone deacetylase 4 has a more favorable binding with Curcumin in comparison to the other histone deacetylases tested.
4. Conclusions
This work is valuable for understanding the specificity of Curcumin towards different Histone deacetylases (HDAC) targets, which can have implications for potential therapeutic applications, especially in the context of diseases associated with HDAC dysregulation.
The findings suggest that Curcumin demonstrates greater affinity for Histone deacetylase 4 compared to other examined histone deacetylases. A more negative binding energy signifies a stronger and more stable interaction, indicating that the binding between Curcumin and Histone deacetylase 4 is more favorable when contrasted with other histone deacetylases that were tested.
Authors Contributions
Protocol designed by IVF . All authors read and approved the final manuscript.
Conflict of Interest
Authors declare that they do not have any conflict of interest.
References
- Marks, P. A., Miller, T., & Richon, V. M. (2003). Histone deacetylases. Current opinion in pharmacology, 3(4), 344-351.
- Gray, S. G., & Ekström, T. J. (2001). The human histone deacetylase family. Experimental cell research, 262(2), 75-83. [CrossRef]
- Barneda-Zahonero, B., & Parra, M. (2012). Histone deacetylases and cancer. Molecular oncology, 6(6), 579-589.
- Cress, W. D., & Seto, E. (2000). Histone deacetylases, transcriptional control, and cancer. Journal of cellular physiology, 184(1), 1-16.
- Marks, P. A., Richon, V. M., Miller, T., & Kelly, W. K. (2004). Histone deacetylase inhibitors. Advances in cancer research, 91, 137-168.
- Glozak, M. A., & Seto, E. (2007). Histone deacetylases and cancer. Oncogene, 26(37), 5420-5432.
- Dokmanovic, M., Clarke, C., & Marks, P. A. (2007). Histone deacetylase inhibitors: overview and perspectives. Molecular cancer research, 5(10), 981-989. [CrossRef]
- Aggarwal, B. B., Kumar, A., & Bharti, A. C. (2003). Anticancer potential of curcumin: preclinical and clinical studies. Anticancer research, 23(1/A), 363-398.
- Vallianou, N. G., Evangelopoulos, A., Schizas, N., & Kazazis, C. (2015). Potential anticancer properties and mechanisms of action of curcumin. Anticancer research, 35(2), 645-651.
- Meng, X. Y., Zhang, H. X., Mezei, M., & Cui, M. (2011). Molecular docking: a powerful approach for structure-based drug discovery. Current computer-aided drug design, 7(2), 146-157. [CrossRef]
- Odhar, H. A., Rayshan, A. M., Ahjel, S. W., Hashim, A. A., & Albeer, A. A. M. A. (2019). Molecular docking enabled updated screening of the matrix protein VP40 from Ebola virus with millions of compounds in the MCULE database for potential inhibitors. Bioinformation, 15(9), 627.
- Wang, A. H., Kruhlak, M. J., Wu, J., Bertos, N. R., Vezmar, M., Posner, B. I., ... & Yang, X. J. (2000). Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Molecular and cellular biology, 20(18), 6904-6912. [CrossRef]
- Vega, R. B., Matsuda, K., Oh, J., Barbosa, A. C., Yang, X., Meadows, E., ... & Olson, E. N. (2004). Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell, 119(4), 555-566. [CrossRef]
- Grozinger, C. M., & Schreiber, S. L. (2000). Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proceedings of the National Academy of Sciences, 97(14), 7835-7840. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).