Submitted:
24 November 2023
Posted:
27 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
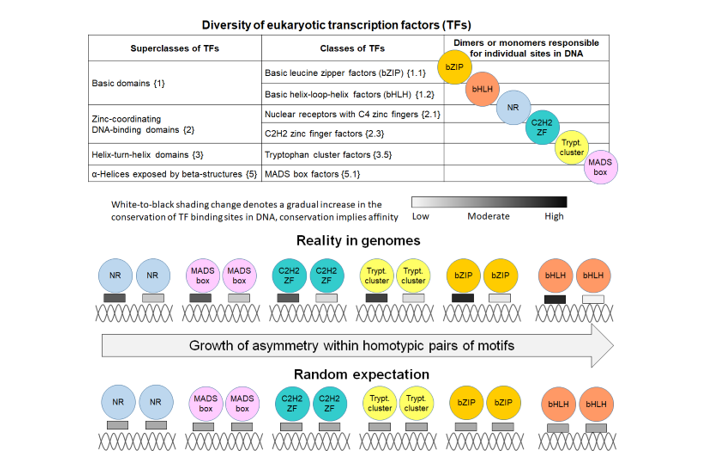
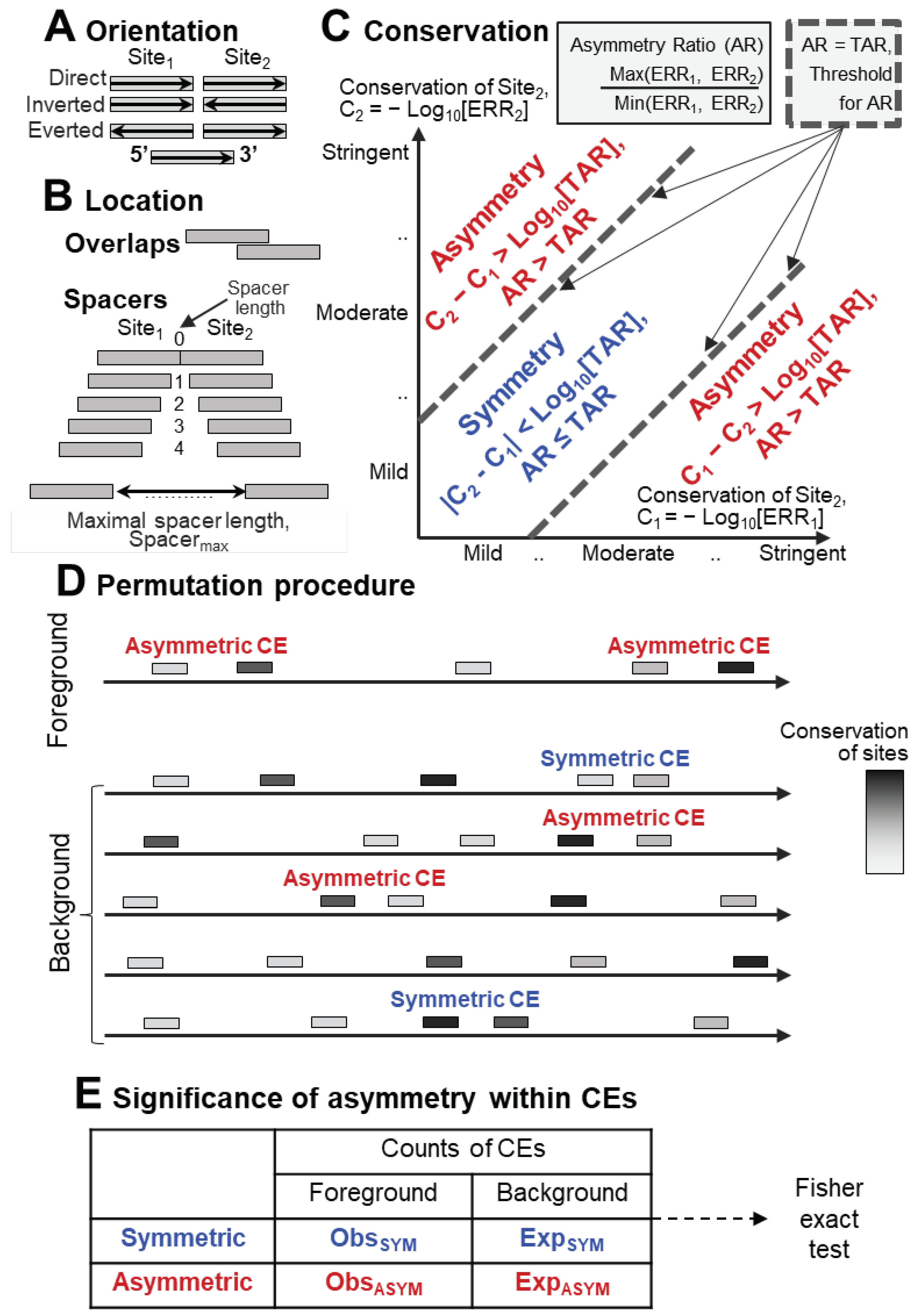
2.1. Definition and examples of asymmetry in the conservation of motifs within CEs
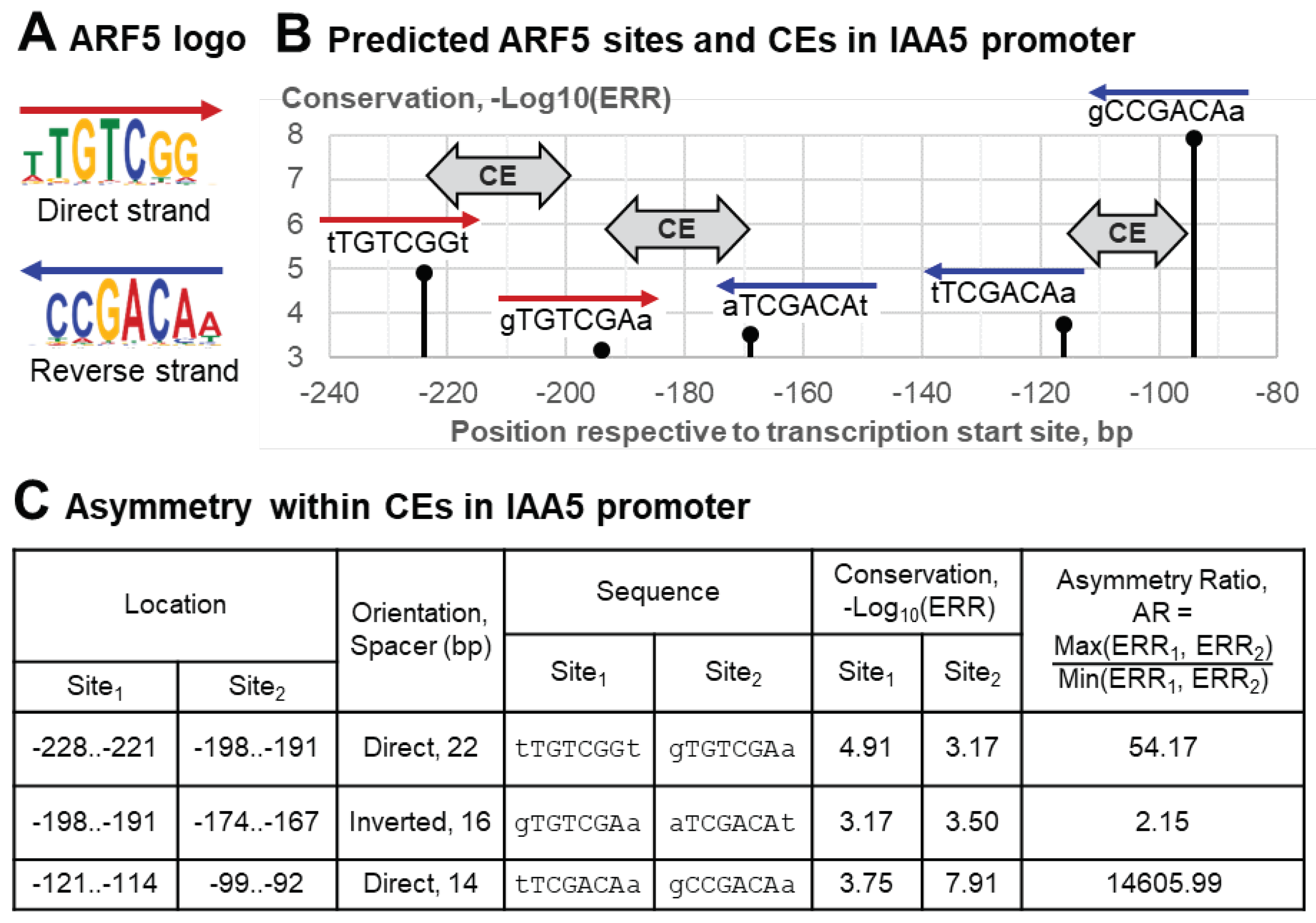
2.2. Analysis of a whole dataset: example of significant asymmetry within CE
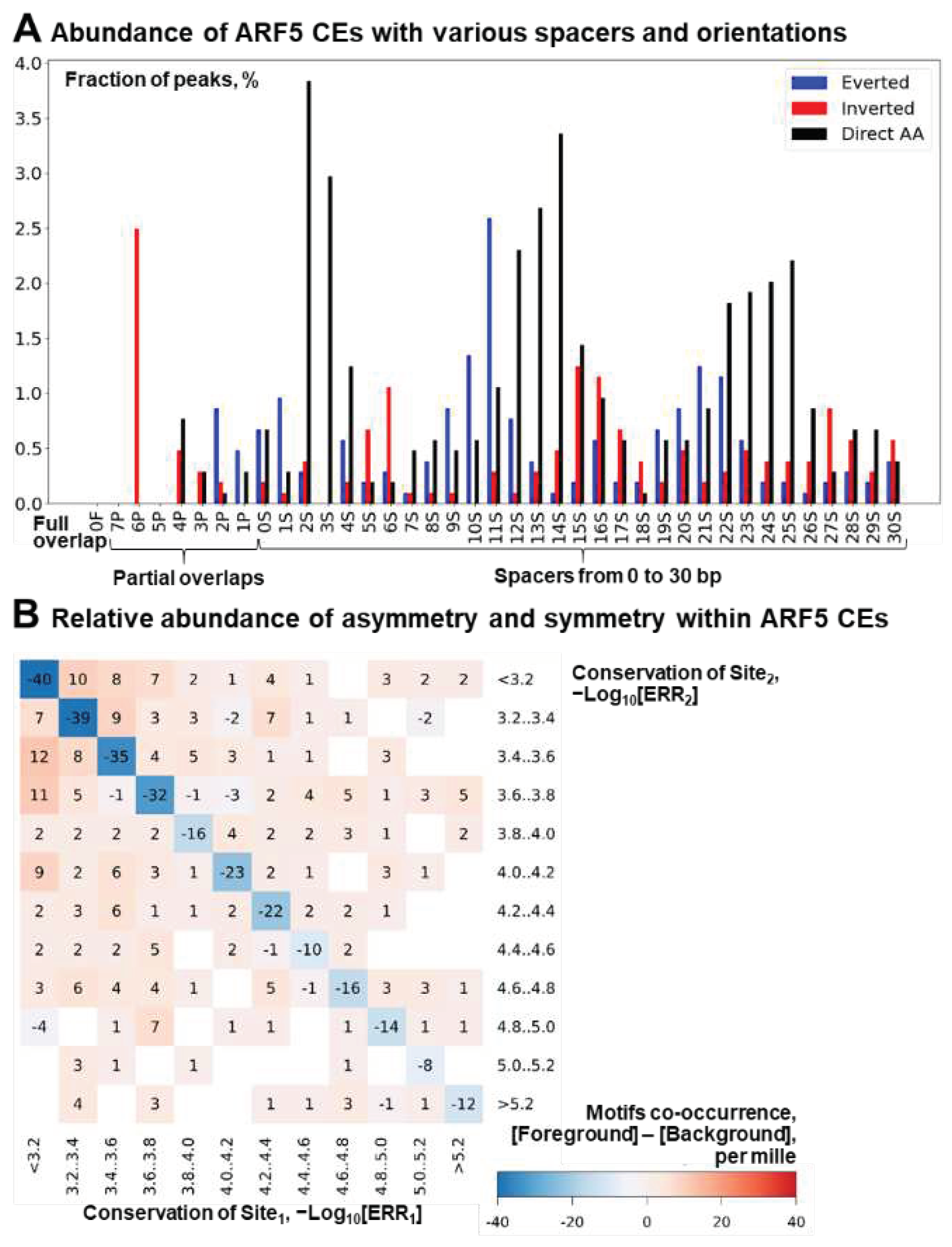
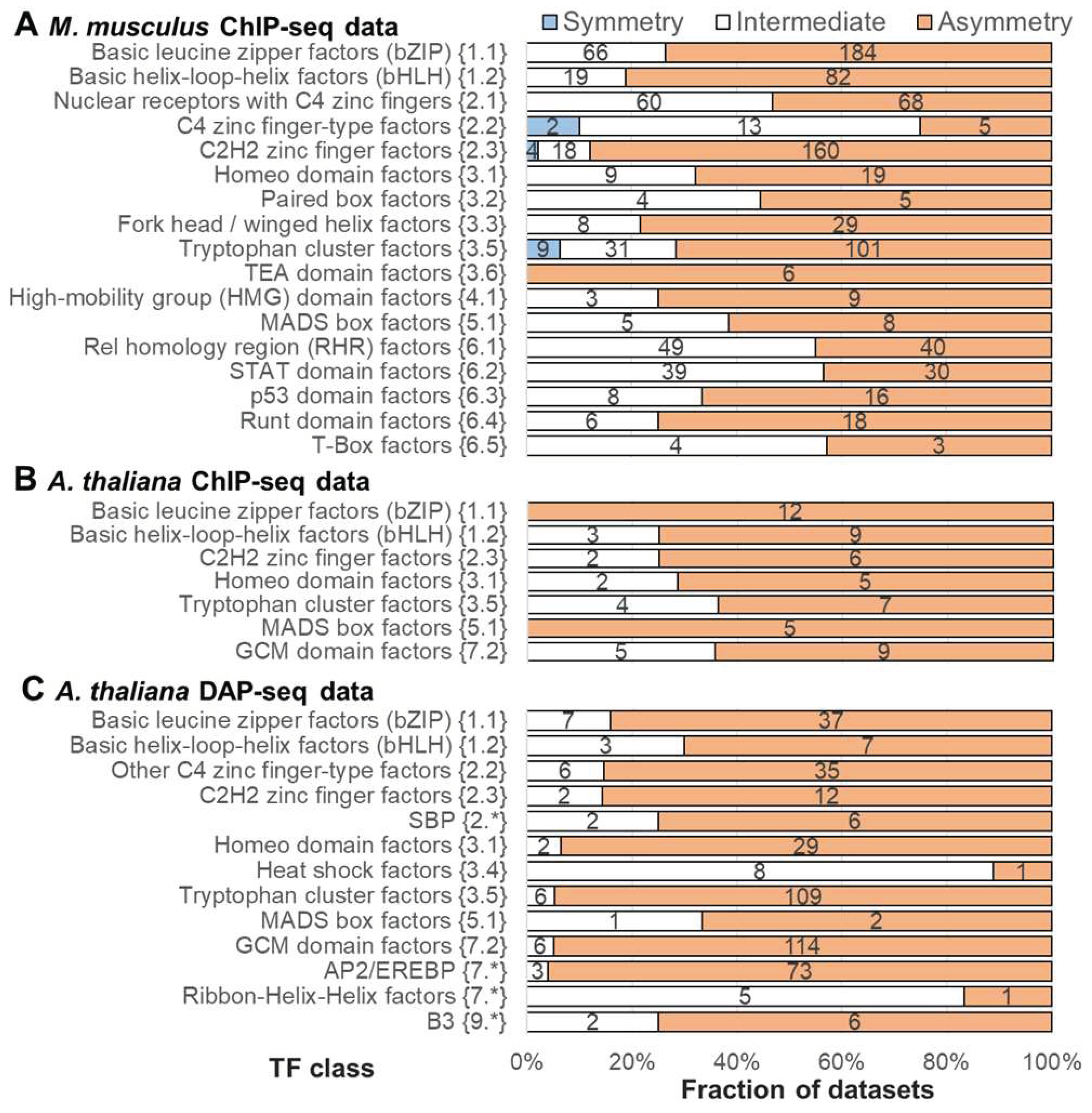
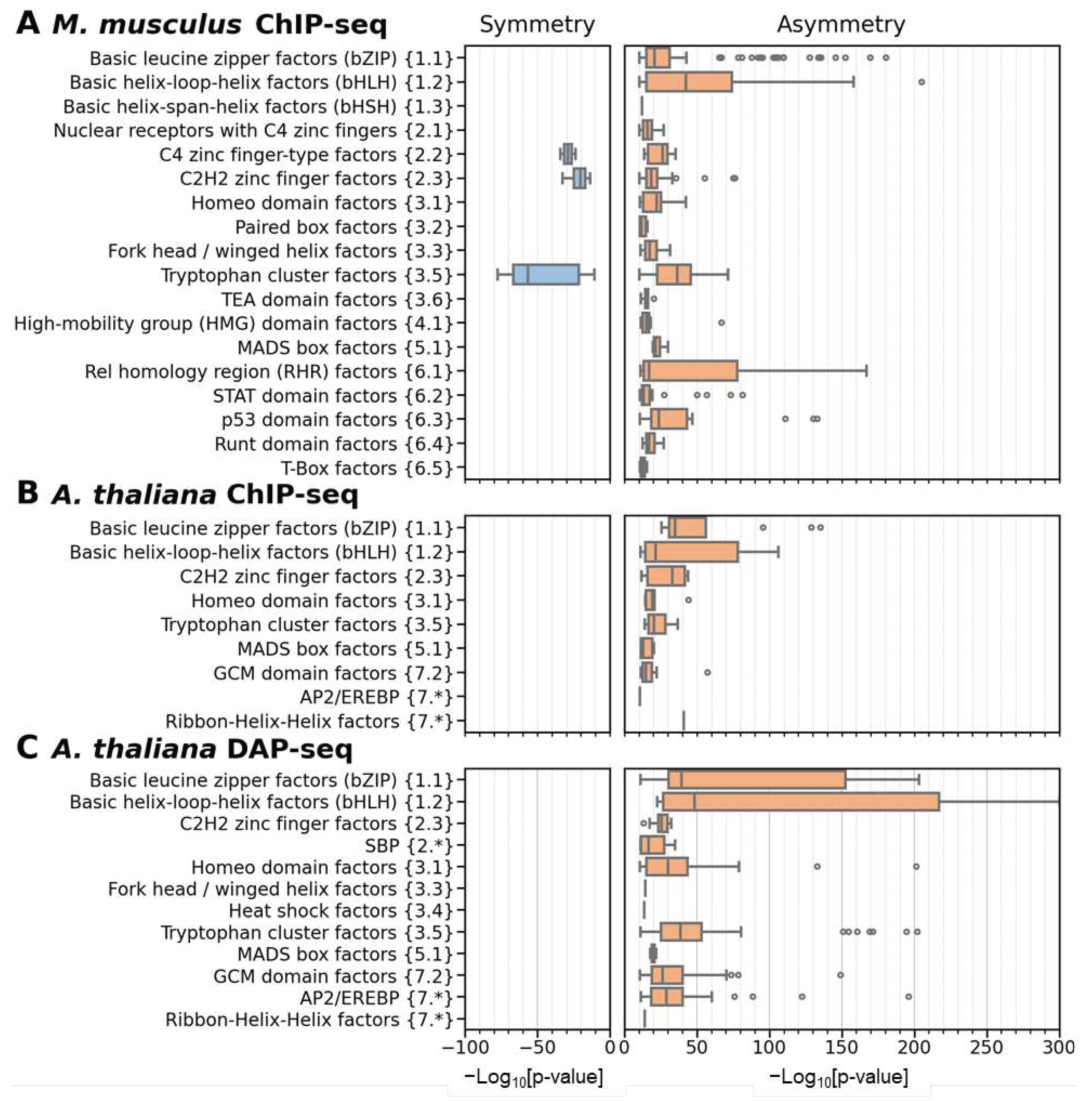
2.3. Massive Analysis of ChIP-seq/DAP-seq reveals that asymmery within homotypic CE depends on the class of target TF
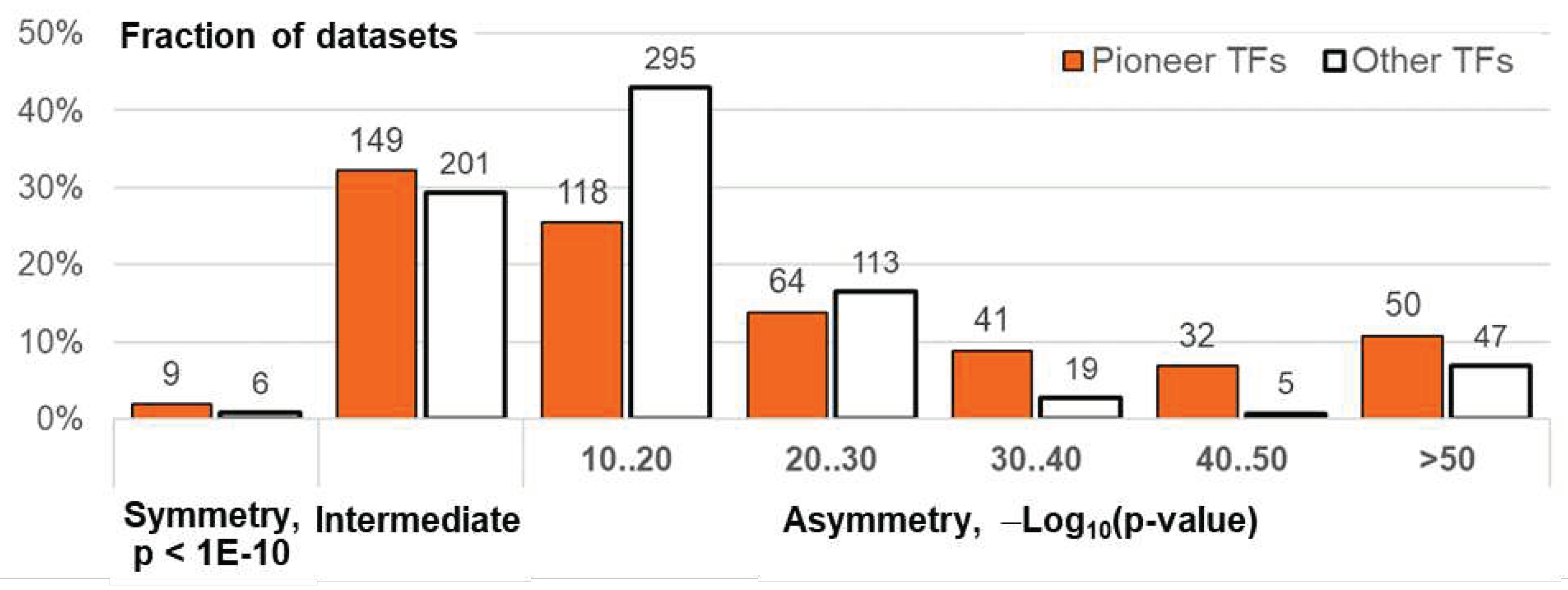
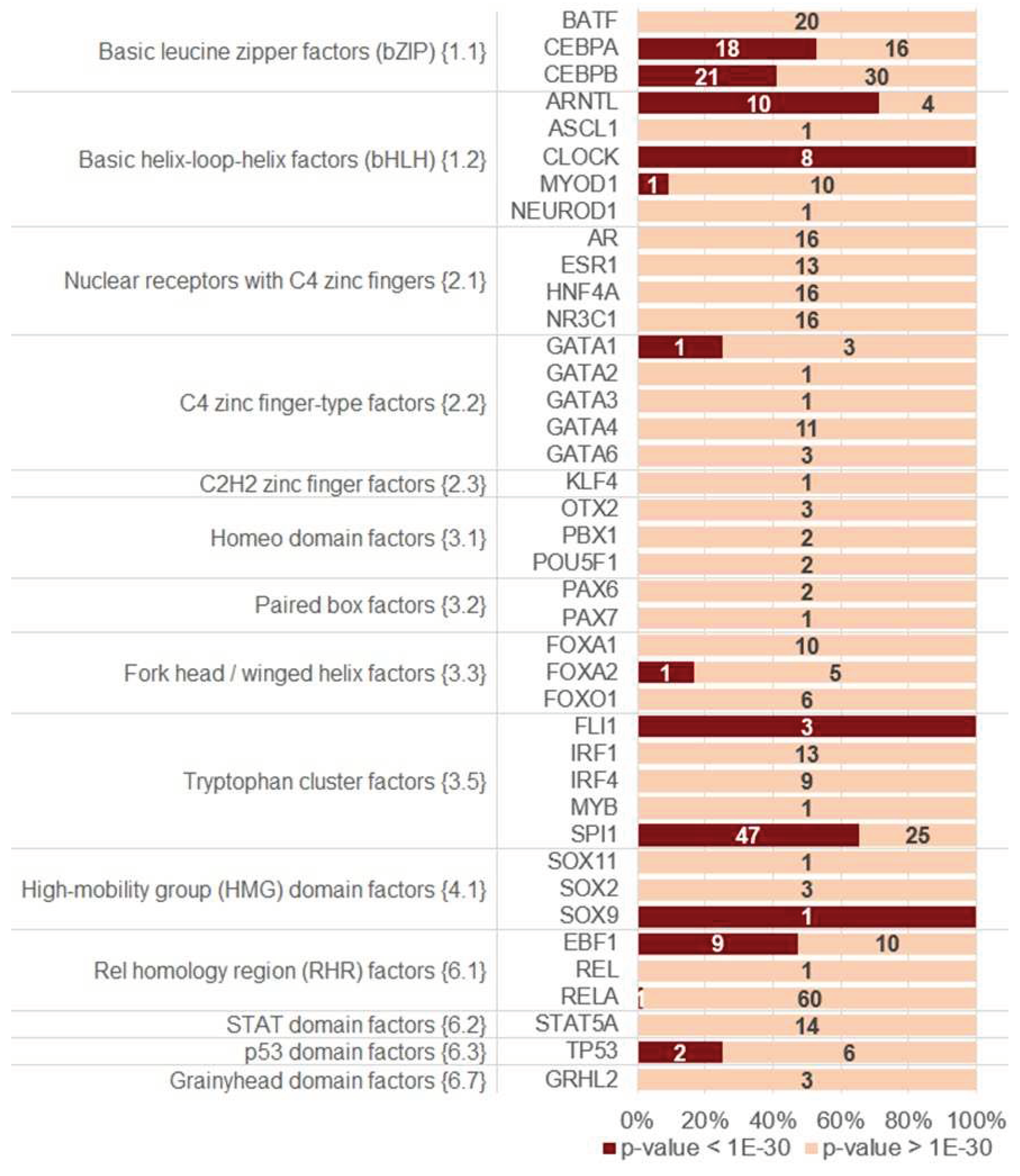
2.3. Homotypic CEs of target TF with proven pioneer activity show higher significance of asymmery compared to that of other TFs
3. Discussion
4. Materials and Methods
4.1. Benchmark collections of ChIP-seq and DAP-seq data, their preliminary filtration
4.2. De novo motif search and final filtration of ChIP-seq/DAP-seq data
4.3. Composite elements analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakato, R.; Shirahige, K. Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation. Brief Bioinform. 2017, 18, 279-290. [CrossRef]
- Lloyd, S.M.; Bao, X. Pinpointing the genomic localizations of chromatin-associated proteins: the yesterday, today, and tomorrow of ChIP-seq. Curr Protoc Cell Biol. 2019, 84, e89. [CrossRef]
- Johnson, D. S.; Mortazavi, A.; Myers, R. M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316(5830), 1497–1502. [CrossRef]
- O'Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 2016, 165, 1280–1292. [CrossRef]
- Zhang, Y.; Li, Z.; Zhang, Y.; Lin, K.; Peng, Y.; Ye, L.; Zhuang, Y.; Wang, M.; Xie, Y.; Guo, J.; et al. Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements. Genome Res. 2021, 31(12), 2276–2289. [CrossRef]
- Wingender, E. Criteria for an updated classification of human transcription factor DNA-binding domains. J Bioinform Comput Biol. 2013, 11(1), 1340007. [CrossRef]
- Wingender, E.; Schoeps, T.; Dönitz, J. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 2013, 41(Database issue), D165–D170. [CrossRef]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Dönitz, J. TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res. 2015, 43(Database issue), D97–D102. [CrossRef]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Krull, M.; Dönitz, J. TFClass: expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018, 46, D343-D347. [CrossRef]
- Castro-Mondragon, J. A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R. B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50(D1), D165–D173. [CrossRef]
- Blanc-Mathieu, R.; Dumas, R.; Turchi, L.; Lucas, J.; Parcy, F. Plant-TFClass: a structural classification for plant transcription factors. Trends Plant Sci. 2023, S1360-1385(23), 00227-3. [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human transcription factors. Cell 2018, 172, 650–665. [CrossRef]
- Morgunova, E.; Taipale, J. Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 2017, 47, 1–8. [CrossRef]
- Reiter, F.; Wienerroither, S., Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017, 43, 73–81. [CrossRef]
- Zeitlinger, J. Seven myths of how transcription factors read the cis-regulatory code. Curr Opin Syst Biol. 2020, 23, 22-31 . [CrossRef]
- Kribelbauer, J. F., Rastogi, C., Bussemaker, H. J., Mann, R. S. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu Rev Cell Dev Biol., 2019, 35, 357–379. [CrossRef]
- Mayran, A.; Drouin, J. Pioneer transcription factors shape the epigenetic landscape. J Biol Chem. 2018, 293, 13795-13804. [CrossRef]
- Bulyk, M. L.; Drouin, J.; Harrison, M. M.; Taipale, J.; Zaret, K. S. Pioneer factors - key regulators of chromatin and gene expression. Nature Rev. Genet. 2023. [CrossRef]
- Lai, X.; Verhage, L.; Hugouvieux, V.; Zubieta, C. Pioneer factors in animals and plants-colonizing chromatin for gene regulation. Molecules 2018, 23, e1914. [CrossRef]
- Zaret, K. S.; Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011, 25, 2227-2241. [CrossRef]
- Whitington, T.; Frith, M.C.; Johnson, J.; Bailey, T.L. Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res. 2011, 39, 98. [CrossRef]
- Guo, Y.; Mahony, S.; Gifford, D.K. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012, 8, e1002638. [CrossRef]
- Kazemian, M.; Pham, H.; Wolfe, S.A.; Brodsky, M. H.; Sinha, S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013, 41, 8237-8352. [CrossRef]
- Jankowski, A.; Prabhakar, S.; Tiuryn, J. TACO: a general-purpose tool for predicting cell-type-specific transcription factor dimers. BMC Genomics 2014, 15, 208. [CrossRef]
- Toivonen, J.; Kivioja, T.; Jolma, A.; Yin, Y.; Taipale, J.; Ukkonen, E. Modular discovery of monomeric and dimeric transcription factor binding motifs for large data sets. Nucleic Acids Res. 2018, 46(8), e44. [CrossRef]
- Levitsky, V.; Zemlyanskaya, E.; Oshchepkov, D.; Podkolodnaya, O.; Ignatieva, E.; Grosse, I.; Mironova, V.; Merkulova, T. A single ChIP-seq dataset is sufficient for comprehensive analysis of motifs co-occurrence with MCOT package. Nucleic Acids Res. 2019, 47, e139. [CrossRef]
- Jolma, A.; Yan, J.; Whitington, T.; Toivonen, J.; Nitta, K. R.; Rastas, P.; Morgunova, E.; Enge, M.; Taipale, M.; Wei, G.; et al. DNA-binding specificities of human transcription factors. Cell 2013, 152, 327–339. [CrossRef]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [CrossRef]
- Kulakovskiy, I.V.; Vorontsov, I.E.; Yevshin, I.S.; Sharipov, R.N.; Fedorova, A.D.; Rumynskiy, E.I.; Medvedeva, Y.A.; Magana-Mora, A.; Bajic, V.B.; Papatsenko, D.A.; et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2018, 46, D252-D259. [CrossRef]
- Ambrosini, G.; Vorontsov, I.; Penzar, D.; Groux, R.; Fornes, O.; Nikolaeva, D. D.; Ballester, B.; Grau, J.; Grosse, I.; Makeev, V.; Kulakovskiy, I.; Bucher, P. Insights gained from a comprehensive all-against-all transcription factor binding motif benchmarking study. Genome Biol 2020, 21, 114. [CrossRef]
- Levitsky, V.; Oshchepkov, D.; Zemlyanskaya, E.; Merkulova, T. Asymmetric conservation within pairs of co-occurred motifs mediates weak direct binding of transcription factors in ChIP-seq data. Int. J. Mol. Sci. 2020, 21, 6023. [CrossRef]
- Levitsky, V.G.; Mukhin, A.M.; Oshchepkov, D.Y.; Zemlyanskaya, E.V.; Lashin, S.A. Web-MCOT Server for Motif Co-Occurrence Search in ChIP-Seq Data. Int. J. Mol. Sci. 2022, 23, 8981. [CrossRef]
- Casey, B. H.; Kollipara, R. K.; Pozo, K.; Johnson, J. E. Intrinsic DNA binding properties demonstrated for lineage-specifying basic helix-loop-helix transcription factors. Genome Res. 2018, 28(4), 484–496. [CrossRef]
- Merkulov, V.M.; Merkulova, T.I. Structural variants of glucocorticoid receptor binding sites and different versions of positive glucocorticoid responsive elements: Analysis of GR-TRRD database. J Steroid Biochem Mol Biol. 2009, 115(1-2), 1-8. [CrossRef]
- Nagy, G.; Nagy, L. Motif grammar: the basis of the language of gene expression. Comput Struct Biotec. 2020, 18, 2026-2032. [CrossRef]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: output control in auxin biology. J Exp Bot. 2018, 69(2), 179-188. [CrossRef]
- Stigliani, A.; Martin-Arevalillo, R.; Lucas, J.; Bessy, A.; Vinos-Poyo, T.; Mironova, V.; Vernoux, T.; Dumas, R.; Parcy, F. Capturing auxin response factors syntax using DNA binding models. Mol Plant 2019, 12(6), 822–832. [CrossRef]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Boer, D. R.; et al., Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. . Proc Natl Acad Sci U S A, 2020, 117(39), 24557–24566. [CrossRef]
- Lavrekha, V. V.; Levitsky, V. G.; Tsukanov, A. V.; Bogomolov, A. G.; Grigorovich, D. A.; Omelyanchuk, N.; Ubogoeva, E. V.; Zemlyanskaya, E. V.; Mironova, V. (2022). CisCross: A gene list enrichment analysis to predict upstream regulators in Arabidopsis thaliana. Front Plant Sci. 2022, 13, 942710. [CrossRef]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [CrossRef]
- Kolmykov, S.; Yevshin, I.; Kulyashov, M.; Sharipov, R.; Kondrakhin, Y.; Makeev, V.J.; Kulakovskiy, I. V.; Kel, A.; Kolpakov, F. GTRD: An integrated view of transcription regulation. Nucleic Acids Res. 2021, 49, D104–D111. [CrossRef]
- McLeay, R.C.; Bailey, T.L. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 2010, 11, 165. [CrossRef]
- Tsukanov, A. V.; Mironova, V. V.; Levitsky, V. G. Motif models proposing independent and interdependent impacts of nucleotides are related to high and low affinity transcription factor binding sites in Arabidopsis. Front Plant Sci. 2022, 13, 938545. [CrossRef]
- Gupta, S.; Stamatoyannopolous, J. A.; Bailey, T. L.; Noble, W. S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [CrossRef]
- Wójcikowska, B.; Belaidi, S.; Robert, H. S. Game of Thrones among AUXIN RESPONSE FACTORs - over thirty years of MONOPTEROS research. J Exp Bot. 2023. [CrossRef]
- Ma, J.; Liu, Y.; Zhou, W.; Zhu, Y.; Dong, A.; Shen, W. H. Histone chaperones play crucial roles in maintenance of stem cell niche during plant root development. Plant J. 2018, 95(1), 86–100. [CrossRef]
- Amoutzias, G. D.; Robertson, D. L.; Van de Peer, Y.; Oliver, S. G. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008, 33(5), 220–229. [CrossRef]
- Gramzow, L.; Ritz, M. S.; Theissen, G. On the origin of MADS-domain transcription factors. Trends Genet. 2010, 26(4), 149–153. [CrossRef]
- Escrivá García, H.; Laudet, V.; Robinson-Rechavi, M. Nuclear receptors are markers of animal genome evolution. J Struct Funct Genomics, 2003, 3(1-4), 177–184. [CrossRef]
- Wang, Y.; Levy, D. E. Comparative evolutionary genomics of the STAT family of transcription factors. JAK-STAT, 2012, 1(1), 23–33. [CrossRef]
- Żyła, N.; Babula-Skowrońska, D. Evolutionary consequences of functional and regulatory divergence of HD-Zip I transcription factors as a source of diversity in protein interaction networks in plants. J Mol Evol. 2023. [CrossRef]
- Kohler, J. J.; Metallo, S. J.; Schneider, T. L.; Schepartz, A. DNA specificity enhanced by sequential binding of protein monomers. Proc Natl Acad Sci U S A, 1999, 96(21), 11735–11739. [CrossRef]
- Metallo, S.J.; Schepartz, A. Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat Struct Biol. 1997, 4(2), 115-117. [CrossRef]
- Ecevit, O.; Khan, M. A.; Goss, D. J. Kinetic analysis of the interaction of b/HLH/Z transcription factors Myc, Max, and Mad with cognate DNA. Biochemistry 2010, 49(12), 2627–2635. [CrossRef]
- Holmbeck, S.M.; Dyson, H.J.; Wright, P.E. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J Mol Biol. 1998, 284, 533–539. [CrossRef]
- Tiwari, M.; Oasa, S.; Yamamoto, J.; Mikuni, S.; Kinjo, M. A quantitative study of internal and external interactions of homodimeric glucocorticoid receptor using fluorescence cross-correlation spectroscopy in a live cell. Sci Rep. 2017, 7(1), 4336. [CrossRef]
- Castellanos, M.; Mothi, N.; Muñoz, V. Eukaryotic transcription factors can track and control their target genes using DNA antennas. Nature comm. 2020, 11(1), 540. [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C. A.; Eeckhoute, J.; Johnson, D. S.; Bernstein, B. E.; Nusbaum, C.; Myers, R. M.; Brown, M.; Li, W.; Liu, X. S. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9(9), R137. [CrossRef]
- Tian, F.; Yang, D. C.; Meng, Y. Q.; Jin, J.; Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48(D1), D1104–D1113. [CrossRef]
- Lamesch, P.; Berardini, T. Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D. L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012, 40(Database issue), D1202–D1210. [CrossRef]
- MCOT. Available online: https://github.com/academiq/mcot-kernel (accessed on 30 October 2023).
- WebMCOT. Available online: https://webmcot.sysbio.cytogen.ru/ (accessed on 30 October 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




