1. Introduction
It is well-recognized that RNA is a versatile molecule capable of performing various functions, including storing genetic information, catalyzing chemical reactions, regulating gene expression, and even self-replicating [
1]. RNA molecules can fulfill their functions as stand-alone molecules (mRNA, tRNA), in complexes with proteins (rRNA), or as structural elements integrated within another RNA molecule (riboswitches) [
2]. For most types of RNA molecules, the ability to form specific secondary structure, and subsequently, tertiary structure, plays a crucial role in performing their functions [
3]. Recent advances in experimental and computational methods for detecting RNA secondary structures have unveiled fascinating examples of molecular processes involving RNA molecules, with many studies being associated with RNA viruses, including HIV, influenza, dengue, Zika, and SARS/SARS-CoV-2 viruses [
4].
RNA viruses pose a serious threat to public health and have significant pandemic potential, as evidenced by the recent COVID-19 pandemic caused by the SARS-CoV-2 RNA virus [
5,
6]. RNA viruses have a higher mutation rate compared to DNA viruses, are typically highly contagious, and, in many cases, effective vaccines and treatments for RNA viruses do not exist [
7]. Exploring the intricacies of RNA virus life cycles and the role of their genome’s secondary structure is essential for our ability to manage and control these viruses. The significance of viral RNA secondary structure has been demonstrated at various stages of the viral life cycle, including virus replication, protein synthesis, packaging, evasion of the host immune system, and the hijacking of host cellular factors [
8].
Recent studies have identified numerous important functional structural elements within viral RNA that interact with proteins, other RNA molecules, or small ligands. Thus, it was found that the secondary structure of HIV-1 Rev response element (RRE) provides the basis for selection of HIV-1 mRNA by Rev protein for nucleocytoplasmic transport [
9]. Another example of an important RNA element with an established secondary structure is the SARS-CoV-2 programmed ribosomal frameshifting stimulation element (FSE) [
10], which induces a one-nucleotide backward shift of the ribosome into an overlapping reading frame at a specific frequency. This shift enables the ribosome to bypass a stop codon and translate ORF1b containing five additional proteins. One more example is the flavivirus cis-acting 5′-flanking element (UFS), characterized by a hairpin secondary structure with a U-rich stem. This element regulates the recruitment of the flavivirus replicase through genome cyclization [
11].
It has come to light that the secondary structure also plays a role in host-mediated RNA editing of RNA viruses, thereby influencing the viral life cycle and the direction of viral evolution. Two families of enzymes—adenosine deaminases (ADAR) and apolipoprotein B mRNA editing catalytic polypeptide family (APOBEC)—were implicated in this process [
12,
13]. The ADAR family members (ADAR1-3) deaminate adenines residing in double-stranded RNA (dsRNA), converting them to inosines (A-to-I) [
14], while APOBEC family members (APOBEC1-2, 3A-H, 4, and AID) deaminate cytosine into uracil (C-to-U) on single-stranded RNA (ssRNA) [
15,
16]. The relative mutational impact between RdRp (RNA-dependent RNA polymerase), which introduces errors during replication, and RNA editing enzymes, remains unclear [
17,
18,
19]. Recent studies revealed an enrichment of C-to-U substitutions and, to a lesser extent, A-to-G substitutions in the SARS-CoV-2 genome, offering evidence for RNA editing by APOBEC and potentially by ADAR enzymes [
20,
21,
22,
23,
24,
25]. However, the full extent of this editing can only be conclusively established through experiments using ADAR and APOBEC knock-out cell lines [
26,
27]. As mentioned earlier, the locations of mutations induced by ADAR and APOBEC depend on secondary structure. Besides APOBEC enzymes’ activity toward single-stranded DNA, Buisson et al. [
28,
29] discovered additional APOBEC preferences that depend on secondary structure. They identified hotspots of APOBEC-induced mutations in cytosines located at the 3′ end of hairpin loops, formed by single-stranded DNA/RNA. A study conducted by Nakata et al. [
30] reported an increased number of C-to-T mutations at the tips of bulge or loop regions within the viral RNA secondary structure. These studies have demonstrated that the secondary structure of the viral genome can significantly influence its evolutionary trajectory [
31].
Recognizing the significance of describing diverse secondary structural patterns within viral genomes, it’s worth noting that the predominant formats used for representing secondary structures in bioinformatics studies remain focused on nucleotide pairing only (e.g., dot-bracket and connectivity table (CT) formats) [
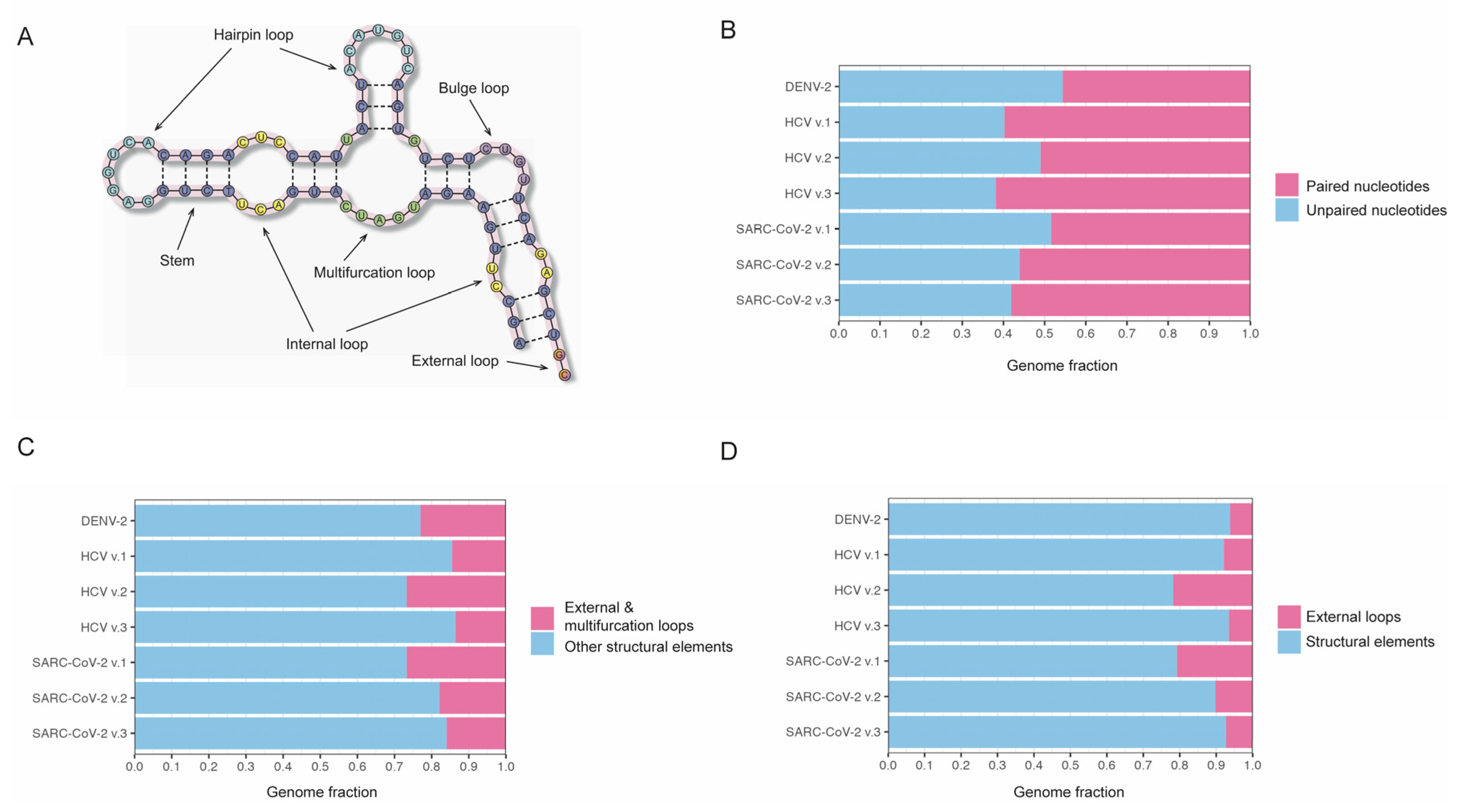
32]. Thus, these formats lack the capability to label higher-order secondary structure elements, such as hairpins, bulges, internal loops, multifurcated loops, and pseudoknots (
Figure 1a). While there is a specialized file format known as Washington University Secondary Structure (WUSS) [
33] designed for describing high-order secondary structure elements, it has not gained common usage thus far. For example, to the best of our knowledge, there are currently no tools available for converting dot-bracket or CT formats to the WUSS format or any packages that offer functionality for conducting a descriptive analysis of secondary structure patterns. To address the gap in this rapidly evolving field, we have developed a Python package for gathering descriptive statistics of high-order secondary structure elements in viral genomes, which also includes the preliminary conversion of conventional secondary structure formats into the WUSS representation. We have also assembled a collection of the currently available secondary structures of viral genomes and used our package to gain insights into the range of secondary structure patterns in these viruses. The package and the collection of secondary structures of viral genomes have been made publicly available for the scientific community.
2. Results
2.1. RNAsselem—a tool for descriptive analysis of high-order RNA secondary structure patterns in viral genomes
The main goals of this study were to develop a programming package for conducting a descriptive analysis of RNA secondary structure patterns in viral genomes and to compile a collection of known secondary structures of RNA viral genomes. The package was designed to perform the following functions: (i) converting from dot-bracket or CT format to WUSS format; (ii) calculating statistics on the pairing and unpairing of nucleotides; (iii) calculating statistics on genome coverage by different types of structural elements; (iv) creating a list of structural elements of a particular type with information on position and size, including hairpin loops, internal loops, bulges, and multifurcation loops; and (v) calculating statistics on structural elements of a given type, providing a frequency of occurrence in the genome, as well as compiting the mean, median, standard deviations of the element size, and the total length of the elements in the genome.
We have also compiled a collection of secondary structure annotations available in the literature for RNA viruses. A total of seven CT files from [
34,
35,
36,
37] were obtained, describing the secondary structures of three RNA viruses, including Dengue virus serotype 2 (DENV-2), Hepatitis C (HCV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Using the developed RNAsselem package, the CT files were converted to a WUSS format, offering a comprehensive annotation of high-order RNA secondary structure patterns. The obtained annotations have been deposited in a GitHub repository to make them freely available to the scientific community. As the WUSS format offers comprehensive annotations for RNA secondary structure elements, we further conducted a detailed descriptive analysis of these structural elements in the collected RNA viruses.
2.2. Abundance of RNA secondary structure elements in the genomes of DENV-2, HCV, and SARS-CoV-2 viruses
Using the developed programming package RNAsselem, we first analyzed the proportion of paired and unpaired nucleotides within the secondary structure of RNA viruses from our dataset (
Figure 1b). Among the secondary structures of viral genomes considered, the DENV-2 virus exhibited the lowest fraction of stem regions (paired nucleotides), accounting for 46%. In comparison, the mean fraction of paired nucleotides in the RNA secondary structures of the HCV virus was 58%, with a standard deviation of 5%, and in the SARS-CoV-2 virus, it was 54% with a standard deviation of 4%. Thus, our findings indicate that approximately half of the nucleotides in the considered RNA viruses are paired.
Next, we analyzed the fractions of the viral genomes occupied by the RNA structural elements. Considering that multifurcation loops are conditionally treated as RNA structural elements, we calculated the fractions of the genomes occupied by RNA elements without (
Figure 1c) and with (
Figure 1d) the inclusion of multifurcation loops. We observed a notable similarity in these fractions across all the analyzed RNA secondary structures of viruses. Thus, the fraction value excluding multifurcation loops for DENV-2 virus, 77%, fell within the error margins of the RNA secondary structure versions of HCV and SARS-CoV-2 viruses, which were 81.7% ± 6% and 79.8% ± 5%, respectively. If multifurcation loops were included in the count of RNA structural elements, the respective proportions of the genome occupied by these elements were 93.7% for DENV, 87.9% ± 7% for HCV, and 87.2% ± 5.7% for SARS-CoV-2. Thus, our observations revealed that a significant portion of the RNA genomes in the studied viruses is characterized by the presence of RNA secondary structure elements.
2.3. Diversity of RNA secondary structure patterns in DENV-2, HCV, and SARS-CoV-2 viruses
We further analyzed the distribution of various RNA structural elements within our collection of the secondary structures of RNA viruses. (
Figure 2 and
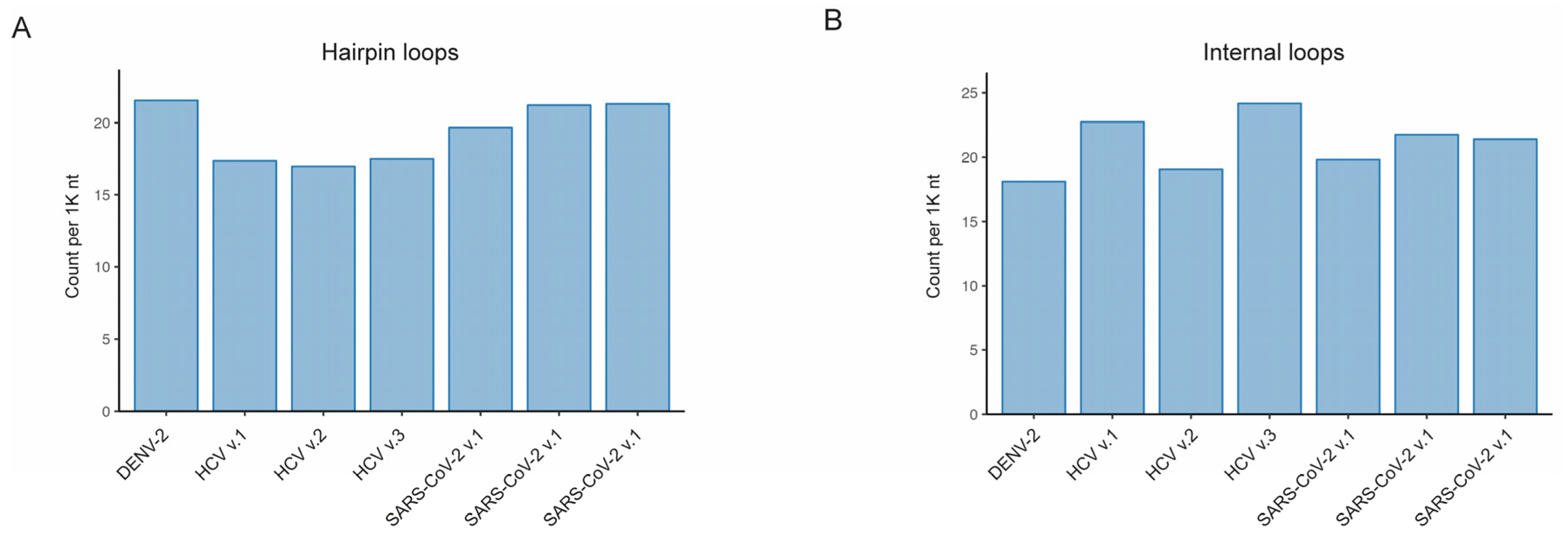
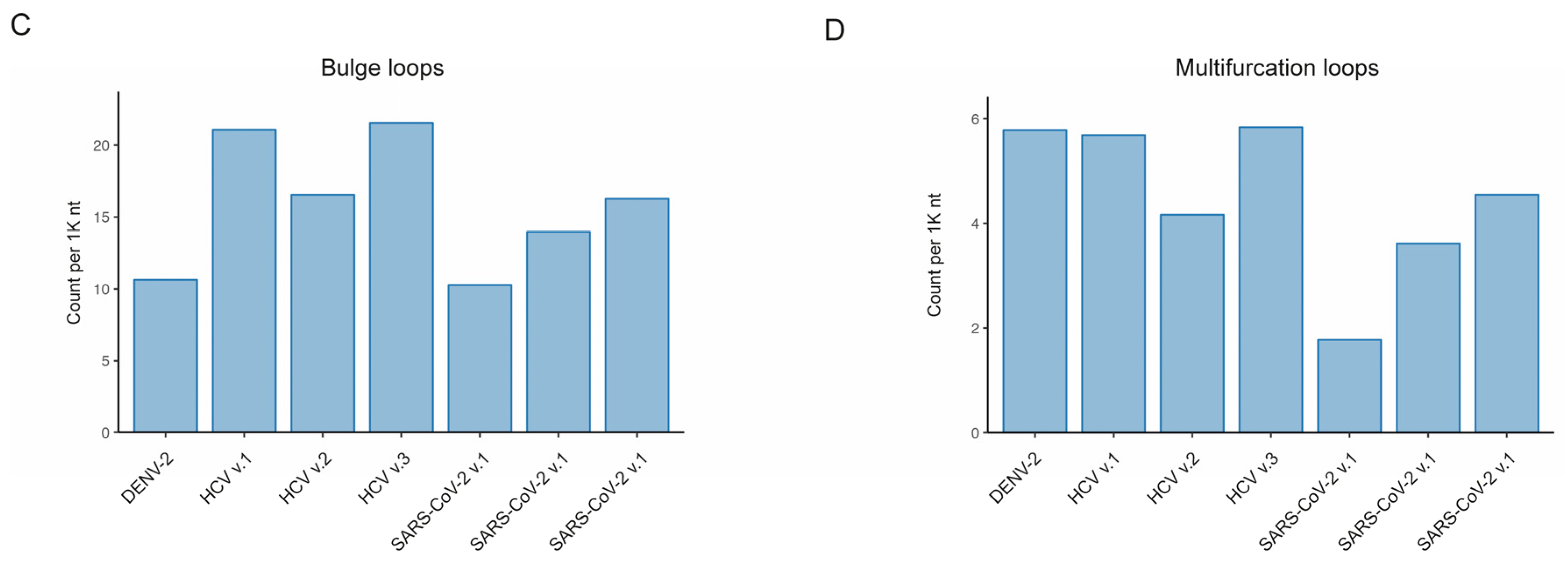
Figure 3). With our developed RNAsselem package, we calculated the number and size of RNA structural elements, such as hairpin loops, internal loops, bulges, and multifurcation loops, for each version of the studied RNA viruses’ secondary structures. The most prevalent RNA structural elements were found to be hairpin loops and internal loops, occurring on average 19.7 and 20.5 times per 1K nucleotides, respectively. The average frequency of bulges was slightly lower at 14.8 occurrences per 1,000 nucleotides. The least frequently observed RNA structural element was multifurcation loops, appearing on average 4.4 times per 1K nt.
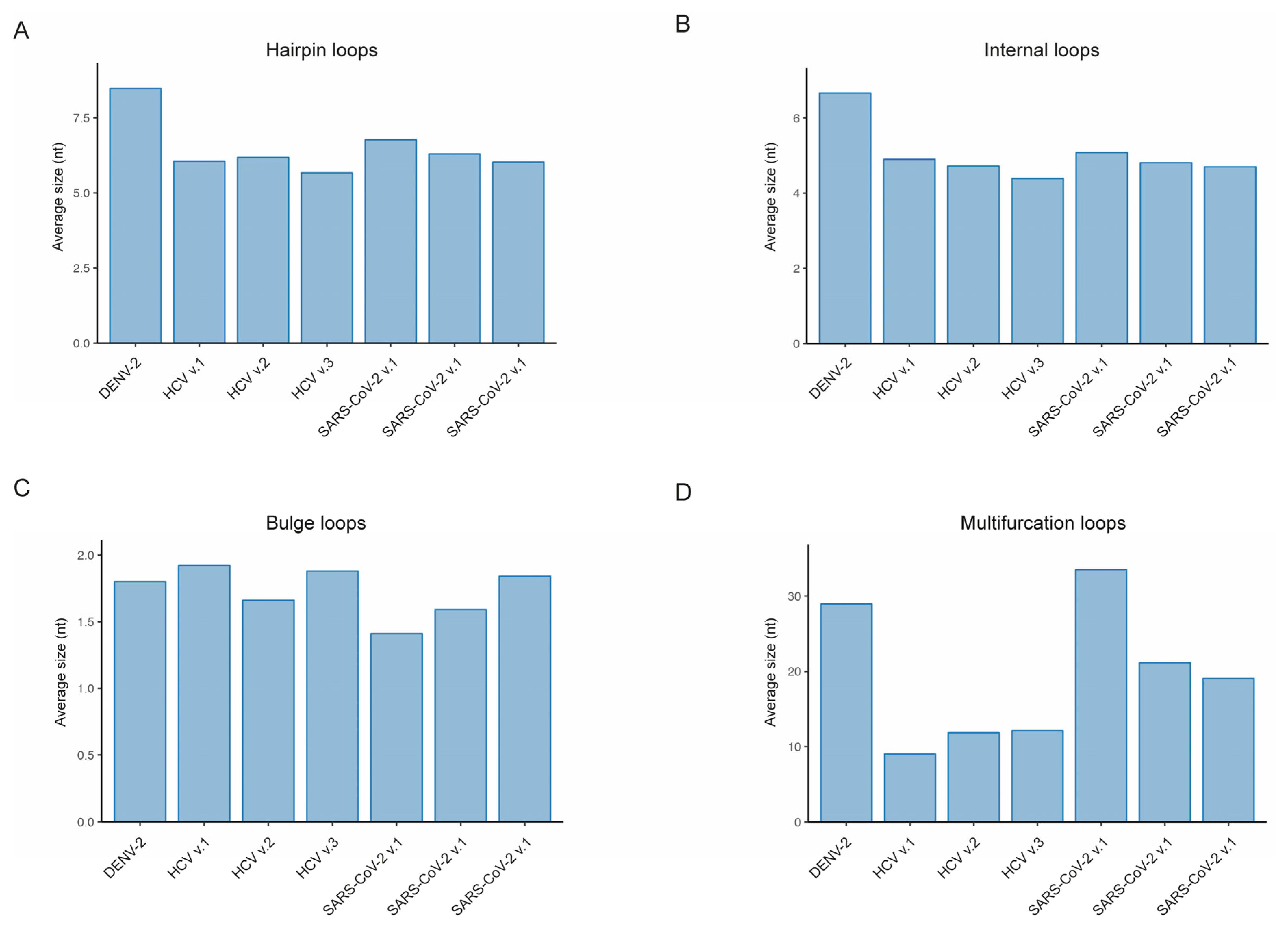
Given the variability in the size of RNA structural elements, we have calculated the total fraction of the genome covered by each type of RNA element for every RNA virus. The highest fraction of nucleotides covered by an RNA structural element was observed for hairpin loops, with an average coverage of 126.7 per 1,000 nucleotides. The second most prominent RNA element, internal loops, demonstrated an average occurrence of 104.8 per 1K nucleotides, and the third most prevalent were multifurcation loops, with an average of 82.3 per 1K nucleotides. The lowest coverage of nucleotides, at 27.8 per 1,000, was observed for bulges. It should be noted that, despite occurring almost as frequently as hairpin loops and internal loops, bulges occupy much less space in RNA viral genomes due to their smaller size.
We have also analyzed variations among the considered viruses in the number and size of RNA elements of the same type. We observed that the frequency of hairpin loops in the HCV virus is slightly lower than in DENV-2 and SARS-CoV-2 viruses: 17.28 ± 0.23 compared to 21.54 and 20.73 ± 0.75, respectively (
Figure 2a). While the frequency of hairpin loops is similar in DENV-2 and SARS-CoV-2, the size, and consequently the coverage, of hairpin loops is higher in the DENV-2 virus compared to SARS-CoV-2: 182.6 versus 131.7 ± 2.4 (
Figure S1a). As observed in
Figure 2b, the frequencies of internal loops are quite similar across the considered viral genomes, while the size of internal loops is higher in the DENV-2 virus compared to other viruses (
Figure 3b): 6.7 versus 4.8 ± 0.2. Due to a higher frequency of bulges in HCV structures, the total coverage was greater in HCV viruses (
Figure S1c), despite the sizes of bulges being approximately similar across all considered genomes (
Figure 3c). The genome coverage by multifurcation loops was similar in HCV and SARS-CoV-2 viruses, as the higher frequency of these structures in HCV (
Figure 2d) was compensated by their larger size in SARS-CoV-2 (
Figure 3d). However, the DENV-2 virus showed significantly higher coverage compared to HCV and SARS-CoV-2, with the number of multifurcation loops similar to those in HCV and a size comparable to the size of these elements in the SARS-CoV-2 virus.
3. Discussion
The increasing evidence highlighting the crucial importance of RNA secondary structure in numerous cellular processes has heightened interest in research within this field. Bioinformatics analysis of RNA structures can offer valuable insights into molecular processes where the structure of the RNA molecule plays a crucial role. Despite the availability of fundamental bioinformatics tools for handling RNA secondary structures, in our opinion, there is a lack of tools for more sophisticated analysis of higher-order RNA secondary structure elements. Here, we introduce a Python package specifically designed for conducting descriptive analyses of RNA secondary structure patterns in genomes of RNA viruses, along with an assembled collection of available secondary structures of viral genomes.
Thus, we have collected available descriptions of RNA secondary structures for the genomes of DENV-2, HCV, and SARS-CoV-2 viruses and applied our package to compare the content of RNA secondary structure elements within these genomes. First, we compared the fractions of the genome occupied by paired nucleotides and observed a similarity across all considered viruses, with approximately half of the genome being covered. Secondly, we found that the fractions of the genome occupied by various RNA structural elements are consistently similar in all genomes, amounting to 80% when excluding multifurcation loops and 90% without exclusion. Third, we compared the statistics for each type of RNA secondary structure element, including the number, size, and genome coverage, and identified variations among viruses. Thus, the hairpin loops, identified as the most common structural RNA element, displayed a larger count and mean size in DENV-2 compared to other viruses. The average size of the internal loops was found to be maximal in the DENV-2 virus. HCV virus surpassed other viruses in the frequency of bulges, while SARS-CoV-2 exhibited a larger size of multifurcation loops, as did the DENV-2 virus, approximately two times bigger than in the HCV virus.
In summary, this study illustrates how our developed bioinformatic package facilitated a comparative descriptive analysis of RNA structural elements across diverse RNA viruses. In general, bioinformatics tools are indispensable for studying the RNA secondary structure in viruses. They enable researchers to analyze and interpret the role of RNA secondary structure, providing insights into its functions and the mechanisms involved in the viral life cycle. Investigating the role of viral RNA secondary structures is crucial for understanding the mechanisms of viral replication and evolution. It could have practical applications in vaccine development and drug design, making it a critical area of research for both basic science and public health.
4. Materials and Methods
4.1. Python package for descriptive analysis of RNA secondary structure elements
The algorithm for converting from the Connectivity Table (CT) format to the Washington University Secondary Structure (WUSS) format was adopted from [
38]. Upon applying this algorithm, paired nucleotides in stem regions were designated using different types of squares (‘<>,’ ‘{},’ ‘[],’ ‘()’) based on their nesting level. Loop regions were classified into the external loops (‘:’), hairpin loops (‘_’), bulges, internal loops (‘-‘), and multifurcation loops (‘,’). The conversion output could be optionally generated in one of two formats: either in WUSS notation or in the CT-like extended format, where the WUSS string is added as an additional column.
The logic of enumerating RNA structural elements was specific for each element type. For hairpin loops and bulges, consecutive labels of particular type were logically concatenated, treating them as a unified RNA element. In the case of internal loops, we interpreted this structural element as two distinct loops: one on the direct strand of the stem and the other on the complementary strand. The combination of these two loops was considered as a single structural element. Similarly, multifurcation labels were initially concatenated into the arcs of multifurcation loops, and then these arcs were organized into the components of multifurcation loops based on the topological analysis of adjusted stems. Stems interrupted by bulges or internal loops were treated as components of a single, integrated stem. A comprehensive overview of the package’s functionality is provided in its documentation. Package documentation and source code are available at:
http://github.com/KazanovLab/RNAsselem.
4.2. Collection of RNA secondary structures of viral genomes
Secondary structures of RNA viruses were retrieved from publications through a PubMed search using the keywords ‘RNA secondary structure’ in combination with the respective RNA virus names. The search was performed for poliovirus, dengue, Zika, coronavirus (specific type, e.g., SARS-CoV-2), hepatitis, and HIV viruses. Among the retrieved publications, we selected those that presented genome secondary structures in dot-bracket or connectivity table (CT) format files. In total, seven CT files describing the structures of three RNA viruses, including Dengue virus serotype 2 (DENV-2), Hepatitis C (HCV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), were obtained. Detailed descriptions of the obtained RNA secondary structures are provided in Supplemental
Table S1. For each CT file in the collected dataset RNA viral secondary structures, we used the developed package to generate both the WUSS-format file and the modified CT-format file, containing an additional column representing the WUSS notation of the secondary structure. The compiled collection is now accessible to the scientific community via the GitHub repository at
http://github.com/KazanovLab/RNAsselem. The repository is organized as follows: secondary structure files of the RNA viruses are located in separate folders, each named according to the short name of the respective virus.
Each folder contains original CT files named according to the virus, concatenated with the PubMed ID of the publication from which these secondary structures were selected. If the original publications contained several secondary structures, the PubMed ID label was concatenated with a suffix explaining the origin of the structure mentioned in the publication. For example, the name of the cell line was included if several cell lines were used in the study. The generated WUSS files and CT-modified files, which include an additional column with WUSS notation, were created with the same names as the original CT files and with extensions ‘
.wuss’
and ‘
.ctwuss’
, respectively.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Author Contributions
Conceptualization, M.D.K.; methodology, M.D.K.; software, F.M.K., G.V.P., E.V.M. and M.D.K.; validation, F.M.K., G.V.P. and E.V.M.; investigation, F.M.K., E.V.M., G.V.P., D.N.I. and M.D.K.; data curation, F.M.K.; writing—original draft preparation, M.D.K.; writing—review and editing, M.D.K.; visualization, E.V.M.; supervision, M.D.K.; project administration, M.D.K.; funding acquisition, D.N.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 22-14-00132.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank Irina Ponomareva for the secondary structure artwork.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Caprara, M.G.; Nilsen, T.W. RNA: Versatility in Form and Function. Nat. Struct. Biol. 2000, 7, 831–833. [CrossRef]
- Strobel, E.J.; Watters, K.E.; Loughrey, D.; Lucks, J.B. RNA Systems Biology: Uniting Functional Discoveries and Structural Tools to Understand Global Roles of RNAs. Curr. Opin. Biotechnol. 2016, 39, 182–191. [CrossRef]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The Roles of Structural Dynamics in the Cellular Functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489. [CrossRef]
- Spitale, R.C.; Incarnato, D. Probing the Dynamic RNA Structurome and Its Functions. Nat. Rev. Genet. 2023, 24, 178–196. [CrossRef]
- Jamison, D.A.; Anand Narayanan, S.; Trovão, N.S.; Guarnieri, J.W.; Topper, M.J.; Moraes-Vieira, P.M.; Zaksas, V.; Singh, K.K.; Wurtele, E.S.; Beheshti, A. A Comprehensive SARS-CoV-2 and COVID-19 Review, Part 1: Intracellular Overdrive for SARS-CoV-2 Infection. Eur. J. Hum. Genet. 2022, 30, 889–898. [CrossRef]
- Narayanan, S.A.; Jamison, D.A.; Guarnieri, J.W.; Zaksas, V.; Topper, M.; Koutnik, A.P.; Park, J.; Clark, K.B.; Enguita, F.J.; Leitão, A.L.; et al. A Comprehensive SARS-CoV-2 and COVID-19 Review, Part 2: Host Extracellular to Systemic Effects of SARS-CoV-2 Infection. Eur. J. Hum. Genet. 2023. [CrossRef]
- Villa, T.G.; Abril, A.G.; Sánchez, S.; de Miguel, T.; Sánchez-Pérez, A. Animal and Human RNA Viruses: Genetic Variability and Ability to Overcome Vaccines. Arch. Microbiol. 2021, 203, 443–464. [CrossRef]
- Boerneke, M.A.; Ehrhardt, J.E.; Weeks, K.M. Physical and Functional Analysis of Viral RNA Genomes by SHAPE. Annu. Rev. Virol. 2019, 6, 93–117. [CrossRef]
- Fang, X.; Wang, J.; O’Carroll, I.P.; Mitchell, M.; Zuo, X.; Wang, Y.; Yu, P.; Liu, Y.; Rausch, J.W.; Dyba, M.A.; et al. XAn Unusual Topological Structure of the HIV-1 Rev Response Element. Cell 2013, 155, 594. [CrossRef]
- Hill, C.H.; Brierley, I. Structural and Functional Insights into Viral Programmed Ribosomal Frameshifting. Annu. Rev. Virol. 2023, 10. [CrossRef]
- Liu, Z.Y.; Li, X.F.; Jiang, T.; Deng, Y.Q.; Ye, Q.; Zhao, H.; Yu, J.Y.; Qin, C.F. Viral RNA Switch Mediates the Dynamic Control of Flavivirus Replicase Recruitment by Genome Cyclization. Elife 2016, 5, 1–27. [CrossRef]
- Kockler, Z.W.; Gordenin, D.A. From RNA World to SARS-CoV-2: The Edited Story of RNA Viral Evolution. Cells 2021, 10, 1557. [CrossRef]
- Zhu, T.; Niu, G.; Zhang, Y.; Chen, M.; Li, C.Y.; Hao, L.; Zhang, Z. Host-Mediated RNA Editing in Viruses. Biol. Direct 2023, 18, 1–12. [CrossRef]
- Piontkivska, H.; Wales-McGrath, B.; Miyamoto, M.; Wayne, M.L. ADAR Editing in Viruses: An Evolutionary Force to Reckon With. Genome Biol. Evol. 2021, 13, 1–21. [CrossRef]
- Klimczak, L.J.; Randall, T.A.; Saini, N.; Li, J.L.; Gordenin, D.A. Similarity between Mutation Spectra in Hypermutated Genomes of Rubella Virus and in SARS-CoV-2 Genomes Accumulated during the COVID-19 Pandemic. PLoS One 2020, 15, 1–21. [CrossRef]
- Kim, K.; Calabrese, P.; Wang, S.; Qin, C.; Rao, Y.; Feng, P.; Chen, X.S. The Roles of APOBEC-Mediated RNA Editing in SARS-CoV-2 Mutations, Replication and Fitness. Sci. Rep. 2022, 12, 1–15. [CrossRef]
- Zong, J.; Zhang, Y.; Guo, F.; Wang, C.; Li, H.; Lin, G.; Jiang, W.; Song, X.; Zhang, X.; Huang, F.; et al. Poor Evidence for Host-Dependent Regular RNA Editing in the Transcriptome of SARS-CoV-2. J. Appl. Genet. 2022, 63, 413–421. [CrossRef]
- Wei, L. Retrospect of the Two-Year Debate: What Fuels the Evolution of SARS-CoV-2: RNA Editing or Replication Error? Curr. Microbiol. 2023, 80, 1–4. [CrossRef]
- Martignano, F.; Di Giorgio, S.; Mattiuz, G.; Conticello, S.G. Commentary on “Poor Evidence for Host-Dependent Regular RNA Editing in the Transcriptome of SARS-CoV-2.” J. Appl. Genet. 2022, 63, 423–428. [CrossRef]
- Di Giorgio, S.; Martignano, F.; Torcia, M.G.; Mattiuz, G.; Conticello, S.G. Evidence for Host-Dependent RNA Editing in the Transcriptome of SARS-CoV-2. Sci. Adv. 2020, 6, 1–9. [CrossRef]
- Simmonds, P.; Azim Ansari, M. Extensive C->U Transition Biases in the Genomes of a Wide Range of Mammalian RNA Viruses; Potential Associations with Transcriptional Mutations, Damage- or Host-Mediated Editing of Viral RNA. PLoS Pathog. 2021, 17, 1–25. [CrossRef]
- Azgari, C.; Kilinc, Z.; Turhan, B.; Circi, D.; Adebali, O. The Mutation Profile of Sars-Cov-2 Is Primarily Shaped by the Host Antiviral Defense. Viruses 2021, 13. [CrossRef]
- Pu, X.; Xu, Q.; Wang, J.; Liu, B. The Continuing Discovery on the Evidence for RNA Editing in SARS-CoV-2. RNA Biol. 2023, 20, 219–222. [CrossRef]
- Liu, X.; Liu, X.; Zhou, J.; Dong, Y.; Jiang, W.; Jiang, W. Rampant C-to-U Deamination Accounts for the Intrinsically High Mutation Rate in SARS-CoV-2 Spike Gene. Rna 2022, 28, 917–926. [CrossRef]
- Wang, J.; Wu, L.; Pu, X.; Liu, B.; Cao, M. Evidence Supporting That C-to-U RNA Editing Is the Major Force That Drives SARS-CoV-2 Evolution. J. Mol. Evol. 2023. [CrossRef]
- Wei, L. Reconciling the Debate on Deamination on Viral RNA. J. Appl. Genet. 2022, 63, 583–585. [CrossRef]
- Cai, H.; Liu, X.; Zheng, X. RNA Editing Detection in SARS-CoV-2 Transcriptome Should Be Different from Traditional SNV Identification. J. Appl. Genet. 2022, 63, 587–594. [CrossRef]
- Buisson, R.; Langenbucher, A.; Bowen, D.; Kwan, E.E.; Benes, C.H.; Zou, L.; Lawrence, M.S. Passenger Hotspot Mutations in Cancer Driven by APOBEC3A and Mesoscale Genomic Features. Science (80-.). 2019, 364. [CrossRef]
- Langenbucher, A.; Bowen, D.; Sakhtemani, R.; Bournique, E.; Wise, J.F.; Zou, L.; Bhagwat, A.S.; Buisson, R.; Lawrence, M.S. An Extended APOBEC3A Mutation Signature in Cancer. Nat. Commun. 2021, 12, 1–11. [CrossRef]
- Nakata, Y.; Ode, H.; Kubota, M.; Kasahara, T.; Matsuoka, K.; Sugimoto, A.; Imahashi, M.; Yokomaku, Y.; Iwatani, Y. Cellular APOBEC3A Deaminase Drives Mutations in the SARS-CoV-2 Genome. Nucleic Acids Res. 2023, 51, 783–795. [CrossRef]
- Ratcliff, J.; Simmonds, P. Potential APOBEC-Mediated RNA Editing of the Genomes of SARS-CoV-2 and Other Coronaviruses and Its Impact on Their Longer Term Evolution. Virology 2021, 556, 62–72. [CrossRef]
- Mathews, D.H. RNA Secondary Structure Analysis Using RNAstructure. Curr. Protoc. Bioinforma. 2014, 1–25. [CrossRef]
- Nawrocki, E.; Eddy, S. RNA Secondary Structures: WUSS Notation, INFERNAL User’s Guide Available online: http://eddylab.org/infernal/Userguide.pdf.
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive Tertiary Structure in the Dengue Virus RNA Genome. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 11513–11518. [CrossRef]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally Conserved Architecture of Hepatitis C Virus RNA Genomes. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 3692–3697. [CrossRef]
- Huston, N.C.; Wan, H.; Strine, M.S.; de Cesaris Araujo Tavares, R.; Wilen, C.B.; Pyle, A.M. Comprehensive in Vivo Secondary Structure of the SARS-CoV-2 Genome Reveals Novel Regulatory Motifs and Mechanisms. Mol. Cell 2021, 81, 584-598.e5. [CrossRef]
- Lan, T.C.T.; Allan, M.F.; Malsick, L.E.; Woo, J.Z.; Zhu, C.; Zhang, F.; Khandwala, S.; Nyeo, S.S.Y.; Sun, Y.; Guo, J.U.; et al. Secondary Structural Ensembles of the SARS-CoV-2 RNA Genome in Infected Cells. Nat. Commun. 2022, 13, 1–14. [CrossRef]
- Eddy, S.; Nawrocki, E.; Carter, N.; Camp, T.; Rivas, E.; Wheeler, T.; Larralde, M.; Horta, D.; Jaenicke, S.; Shands, W.; et al. Easel - a C Library for Biological Sequence Analysis Available online: https://github.com/EddyRivasLab/easel.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).