Submitted:
02 November 2023
Posted:
02 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
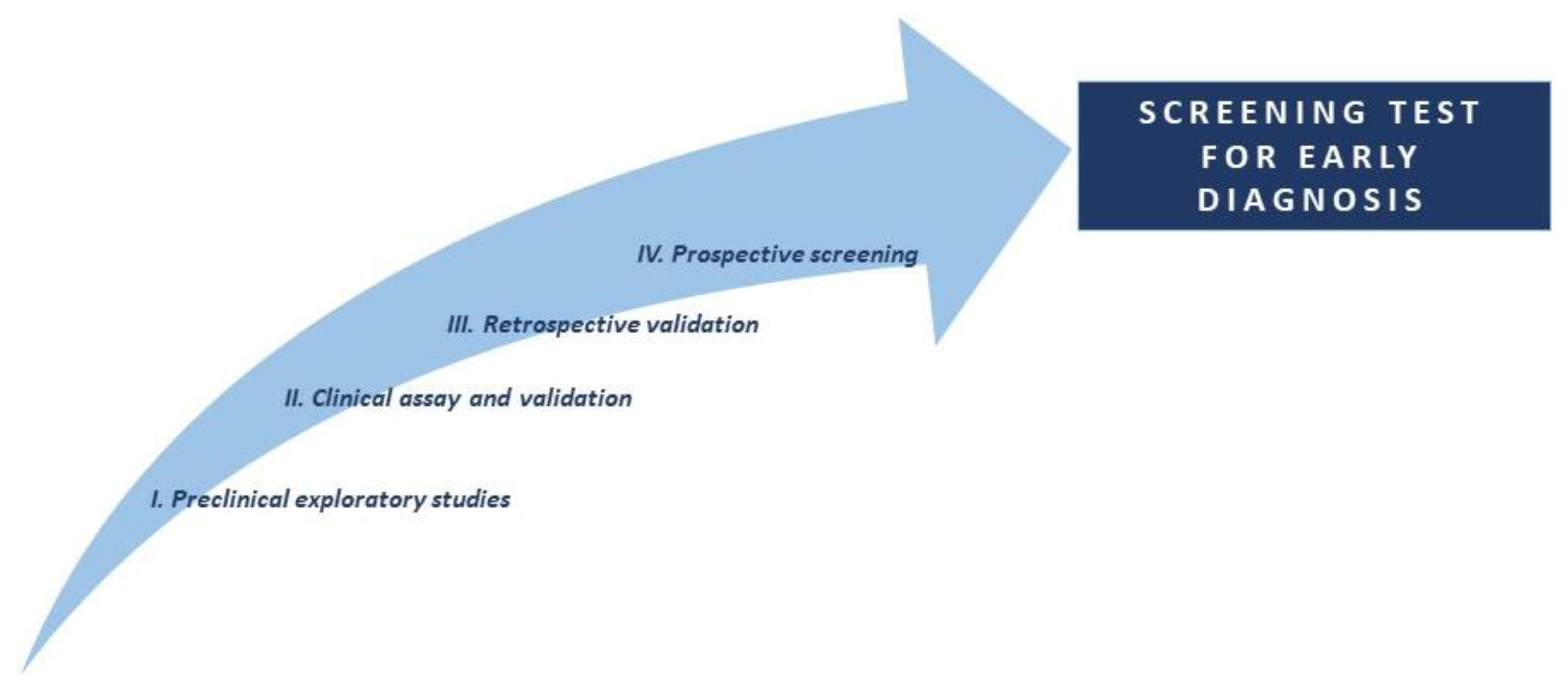
1.1. Phases for the discovery of lung cancer biomarkers
2. Materials and Methods
2.1. Selection of articles
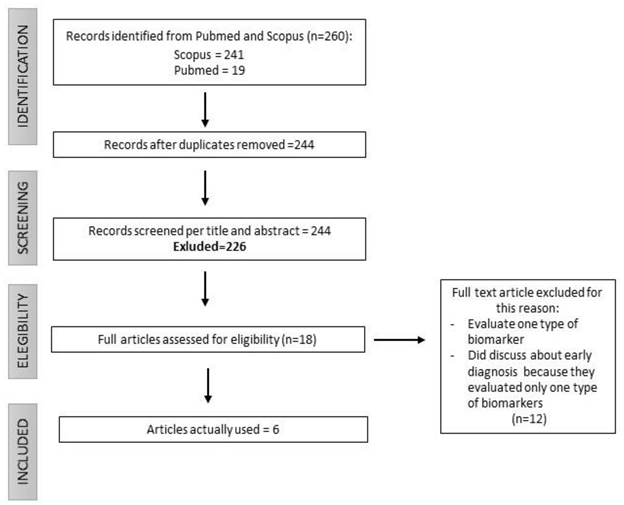
3. Results: Current and Promising Lung Cancer Biomarkers
3.1. Circulating blood proteins and autoantibodies
3.2. microRNA (miRNAs)
3.3. Circulating Tumor Cells (CTCs) and Circulating Tumor DNA (ctDNA)
3.4. Future directions and challenges: Volatile organic compounds (VOCs)
4. Discussion
4.1. Future perspectives
Bibliography
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Leiter, A.; Veluswamy, R.R.; Wisnivesky, J.P. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023, 20, 624–639. [Google Scholar] [CrossRef]
- Wood, D.E.; Kazerooni, E.A.; Aberle, D.; et al. NCCN Guidelines® Insights: Lung Cancer Screening, Version 1.2022. J Natl Compr Canc Netw. 2022, 20, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Dingillo, G.; Bassiri, A.; Badrinathan, A.; et al. Lung Cancer in Young Patients is Associated With More Advanced Disease but Better Overall Survival. J Surg Res. 2023, 292, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Salcedo, P.; Wilson, D.O.; de-Torres, J.P.; et al. Improving selection criteria for lung cancer screening. The potential role of emphysema. Am J Respir Crit Care Med. 2015, 191, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.T.; Wang, P.H.; Chen, W.F.; Lin, C.J. Risk Assessment of Early Lung Cancer with LDCT and Health Examinations. Int J Environ Res Public Health. 2022, 19, 4633. [Google Scholar] [CrossRef] [PubMed]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Saman, H.; Raza, A.; Patil, K.; Uddin, S.; Crnogorac-Jurcevic, T. Non-Invasive Biomarkers for Early Lung Cancer Detection. Cancers (Basel). 2022, 14, 5782. [Google Scholar] [CrossRef]
- Casagrande, G.M.S.; Silva, M.O.; Reis, R.M.; Leal, L.F. Liquid Biopsy for Lung Cancer: Up-to-Date and Perspectives for Screening Programs. Int J Mol Sci. 2023, 24, 2505. [Google Scholar] [CrossRef]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef]
- Xu, B.J.; Gonzalez, A.L.; Kikuchi, T.; et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteomics Clin Appl. 2008, 2, 1508–1517. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Doseeva, V.; Colpitts, T.; Gao, G.; Woodcock, J.; Knezevic, V. Performance of a multiplexed dual analyte immunoassay for the early detection of non-small cell lung cancer. J Transl Med. 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, P.J.; Wang, X.F.; Han, X.; et al. Evaluation of a Serum Lung Cancer Biomarker Panel. Biomark Insights. 2018, 13, 1177271917751608. [Google Scholar] [CrossRef]
- Silvestri, G.A.; Tanner, N.T.; Kearney, P.; et al. Assessment of Plasma Proteomics Biomarker’s Ability to Distinguish Benign From Malignant Lung Nodules: Results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) Trial. Chest. 2018, 154, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Jett, J.R.; Dyer, D.; Kern, J.; Rollins, D.; Phillips, M. Screening for lung cancer with the EarlyCDT-Lung and computed tomography. J Thorac Oncol 2015, 10, S306. [Google Scholar]
- Chapman, C.J.; Healey, G.F.; Murray, A.; et al. EarlyCDT®-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol. 2012, 33, 1319–1326. [Google Scholar] [CrossRef]
- Du, Q.; Yu, R.; Wang, H.; et al. Significance of tumor-associated autoantibodies in the early diagnosis of lung cancer. Clin Respir J. 2018, 12, 2020–2028. [Google Scholar] [CrossRef]
- Paez, R.; Kammer, M.N.; Tanner, N.T.; et al. Update on Biomarkers for the Stratification of Indeterminate Pulmonary Nodules. Chest. 2023, 164, 1028–1041. [Google Scholar] [CrossRef]
- Solassol, J.; Maudelonde, T.; Mange, A.; Pujol, J.L. Clinical relevance of autoantibody detection in lung cancer. J Thorac Oncol. 2011, 6, 955–962. [Google Scholar] [CrossRef]
- Marmor, H.N.; Zorn, J.T.; Deppen, S.A.; Massion, P.P.; Grogan, E.L. Biomarkers in Lung Cancer Screening: a Narrative Review. Curr Chall Thorac Surg. 2023, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Todd, N.W.; Xing, L.; et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010, 127, 2870–2878. [Google Scholar] [CrossRef] [PubMed]
- Le, H.B.; Zhu, W.Y.; Chen, D.D.; et al. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012, 29, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Leidinger, P.; Keller, A.; Backes, C.; Huwer, H.; Meese, E. MicroRNA expression changes after lung cancer resection: a follow-up study. RNA Biol. 2012, 9, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Montani, F.; Marzi, M.J.; Dezi, F.; et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015, 107, djv063. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, U.; Boeri, M.; Sestini, S.; et al. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann Oncol. 2022, 33, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Boeri, M.; Rossi, M.; et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study [published correction appears in J Clin Oncol. 2014, 32, 1520]. J Clin Oncol. 2014, 32, 768–773. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Long-Mira, E.; et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014, 9, e111597. [Google Scholar] [CrossRef]
- Chang, L.; Li, J.; Zhang, R. Liquid biopsy for early diagnosis of non-small cell lung carcinoma: recent research and detection technologies. Biochim Biophys Acta Rev Cancer. 2022, 1877, 188729. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, Z.; Dong, J.; et al. Folate receptor-positive circulating tumor cells as a novel diagnostic biomarker in non-small cell lung cancer. Transl Oncol. 2013, 6, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Katz, R.L.; Zaidi, T.M.; Pujara, D.; et al. Identification of circulating tumor cells using 4-color fluorescence in situ hybridization: Validation of a noninvasive aid for ruling out lung cancer in patients with low-dose computed tomography-detected lung nodules. Cancer Cytopathol. 2020, 128, 553–562. [Google Scholar] [CrossRef]
- Lei, Y.; Sun, N.; Zhang, G.; et al. Combined detection of aneuploid circulating tumor-derived endothelial cells and circulating tumor cells may improve diagnosis of early stage non-small-cell lung cancer. Clin Transl Med. 2020, 10, e128. [Google Scholar] [CrossRef] [PubMed]
- Herath, S.; Sadeghi Rad, H.; Radfar, P.; et al. The Role of Circulating Biomarkers in Lung Cancer. Front Oncol. 2022, 11, 801269. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Ponomaryova, A.A.; Rykova, E.Y.; Cherdyntseva, N.V.; et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013, 81, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lozano Sinues, P.; Zenobi, R.; Kohler, M. Analysis of the exhalome: a diagnostic tool of the future. Chest. 2013, 144, 746–749. [Google Scholar] [CrossRef]
- Schmidt, F.; Kohlbrenner, D.; Malesevic, S.; et al. Mapping the landscape of lung cancer breath analysis: A scoping review (ELCABA). Lung Cancer. 2023, 175, 131–140. [Google Scholar] [CrossRef]
- Chen, X.; Muhammad, K.G.; Madeeha, C.; et al. Calculated indices of volatile organic compounds (VOCs) in exhalation for lung cancer screening and early detection. Lung Cancer. 2021, 154, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Rudnicka, J.; Kowalkowski, T.; Buszewski, B. Searching for selected VOCs in human breath samples as potential markers of lung cancer. Lung Cancer. 2019, 135, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Peled, N.; Hakim, M.; Bunn, P.A. . Jr.; et al. Non-invasive breath analysis of pulmonary nodules. J Thorac Oncol. 2012, 7, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, W.; He, Z.; et al. Detection of lung cancer with electronic nose using a novel ensemble learning framework. J Breath Res. 2021, 15. [Google Scholar] [CrossRef] [PubMed]
- Shlomi, D.; Abud, M.; Liran, O.; et al. Detection of Lung Cancer and EGFR Mutation by Electronic Nose System. J Thorac Oncol. 2017, 12, 1544–1551. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, A.; Beigi, P.; Srinidhi, A.; Lam, S.; MacAulay, C.E. Sex and Smoking Status Effects on the Early Detection of Early Lung Cancer in High-Risk Smokers Using an Electronic Nose. IEEE Trans Biomed Eng. 2015, 62, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Gasparri, R.; Santonico, M.; Valentini, C.; et al. Volatile signature for the early diagnosis of lung cancer. J Breath Res. 2016, 10, 016007. [Google Scholar] [CrossRef]
- Diagnosing Non-Small Cell Lung Cancer by Exhaled Breath Profiling Using an Electronic Nose: A Multicenter Validation Study. Chest. 2023, 163, 697–706. [CrossRef] [PubMed]
- Hanai, Y.; Shimono, K.; Matsumura, K.; et al. Urinary volatile compounds as biomarkers for lung cancer. Biosci Biotechnol Biochem. 2012, 76, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Gasparri, R.; Capuano, R.; Guaglio, A.; et al. Volatolomic urinary profile analysis for diagnosis of the early stage of lung cancer. J Breath Res. 2022, 16. [Google Scholar] [CrossRef]
- Ning, J.; Ge, T.; Jiang, M.; et al. Early diagnosis of lung cancer: which is the optimal choice? Aging (Albany NY). 2021, 13, 6214–6227. [Google Scholar] [CrossRef] [PubMed]
- Otano, I.; Ucero, A.C.; Zugazagoitia, J.; Paz-Ares, L. At the crossroads of immunotherapy for oncogene-addicted subsets of NSCLC. Nat Rev Clin Oncol. 2023, 20, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, M.S.; Ahmed, M. Enzyme-Linked Immunosorbent Assay (ELISA). Methods Mol Biol. 2022, 2508, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Lubin, R.; Zalcman, G.; Bouchet, L.; et al. Serum p53 antibodies as early markers of lung cancer. Nat Med. 1995, 1, 701–702. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




