1. Introduction
Methylation diagnostic testing may seem like a new approach to most clinicians today, although the idea has been around for a long time. DNA methylation in human papillomavirus (HPV) genomes was recognized in the early 1980’s and postulated to have a regulatory role [
1]. The late Professor Harald zur Hausen and his team showed in 1988 that DNA methyltransferase had an important regulatory role in the life cycle of HPV18 [
2]. Methylation as a target for cervical cancer prevention has been actively studied for at least 15 years and is starting to mature into a tool for routine clinical use. New approaches in healthcare can take decades to be incorporated into practice patterns, something that was true for HPV testing, which was already well known as a promising diagnostic target within expert scientific circles in the early 1980s [
3,
4,
5,
6], in fact Dr zur Hausen received his Nobel Prize for research on HPV that was done in the 1970s and early 1980s. General acceptance of HPV as a central part of cervical carcinogenesis took more than 25 years and is today regarded as a key target for standard of care approaches in some countries [
7,
8,
9,
10,
11,
12,
13,
14,
15]. Regrettably, effective tools for control of HPV remain unattainable to most of the world’s population. Dissemination of important new technologies in medicine can be very slow and requires the alignment of scientific, financial, medical, and societal incentives. DNA methylation testing may seem like just another method for finding cervical precancerous lesions and some may ask if there is not already a sufficiently large and confusing number of options for triage? with myriad algorithms and triage variations promoted by different groups and medical bodies. In fact, major problems remain to be solved in cervical cancer screening which include: 1) improved access to the best methods; 2) decreased costs to allow more women to be screened; 3) simplification and shortening of disease diagnosis and management algorithms; 4) Reduction in overtreatment, especially in women of childbearing age; 5) Addressing evolving diagnostic needs as HPV vaccination becomes more widespread.
2. Human Papillomavirus Infection has Many Confusing Manifestations
Efficient detection and management of precancerous cervical disease is hampered by the high prevalence and myriad manifestations of HPV infection. Most HPV infections do not have any detectable cytological expression and can be defined only by molecular methods [
8,
9,
16,
17]. Furthermore, certain tangential cytological appearances of HPV infection, such as atypical squamous cells of undetermined significance (ASCUS) have, as the name implies, very poor specificity [
13,
15]. A diagnosis of low grade squamous intraepithelial lesion (LSIL) is usually but not always associated with HPV infection but almost always these lesions are transient with a low risk of progression to precancer and cancer. The genotype of HPV is a key determinant of risk for invasive cancer and this characteristic cannot be determined by cytology. The proportion of generally healthy women in a population who have detectable borderline or mildly abnormal changes in their cytology (for example ASCUS or LSIL), or are infected by high risk human papillomavirus (hrHPV) is large, typically at between 10 to 20% [
9,
10,
11,
12,
13,
14,
15,
16,
17]. A substantial majority of these women are not at risk for cervical cancer but many become caught in a diagnostic nightmare. Current algorithms and cervical disease management protocols are complex and problematic. Women are frequently demotivated by screening and surveillance methods that are invasive, repetitive, and may stretch over decades; such scenarios produce emotional anxiety, loss to followup and a general unwillingness to engage in screening activities [
15,
16].
3. HPV Persistence and Elevated Methylation
Persistence of hrHPV infection is the major easily measurable risk factor for cervical cancer but it requires at least three HPV genotyping tests over a period of up to two years or longer and depends on the rather imprecise definition of persistence. On rare occasions an invasive cancer may develop after just a few years of infection but more typically there is an extensive lag period of 10 to 20+ years before the invasive cancer emerges [
9,
16]. Cytology has been seen as a key part of cervical cancer prevention for more than 50 years but confidence in cytology has been waning. The general recognition that hrHPV testing detects more precancers and cancers than cytology and that negative HPV results are much more reassuring for low risk of developing cancer than negative cytology has led to an increasing use of hrHPV tests in screening [
14,
16,
18]. A limitation of HPV testing is a moderate to poor specificity in detecting precancers, including cervical intraepithelial neoplasia grades 2 or 3 (CIN2/3), usually less than 90%; which may seem superficially acceptable but needs to be considered in the context of the relatively low prevalence of CIN3 and cervical cancer in most populations. Low specificity plus low disease prevalence means low positive predictive value (PPV), which leads to overtreatment of incorrectly assumed cervical cancer precursors, a situation that motivates the need for better triage before referral of women to colposcopy. Cytology and HPV16/18 genotyping are the common triage tests in current use but positive results on these tests still allow 75% - 90% of women going through colposcopy clinics to not have any treatable CIN2/3 [
15,
16,
17]. There is a need for better triage tests to decrease colposcopy rates, which would also permit clinicians to have improved focus on the women who really have precancers.
4. Characteristics of DNA Methylation and Relationship to HPV Persistence
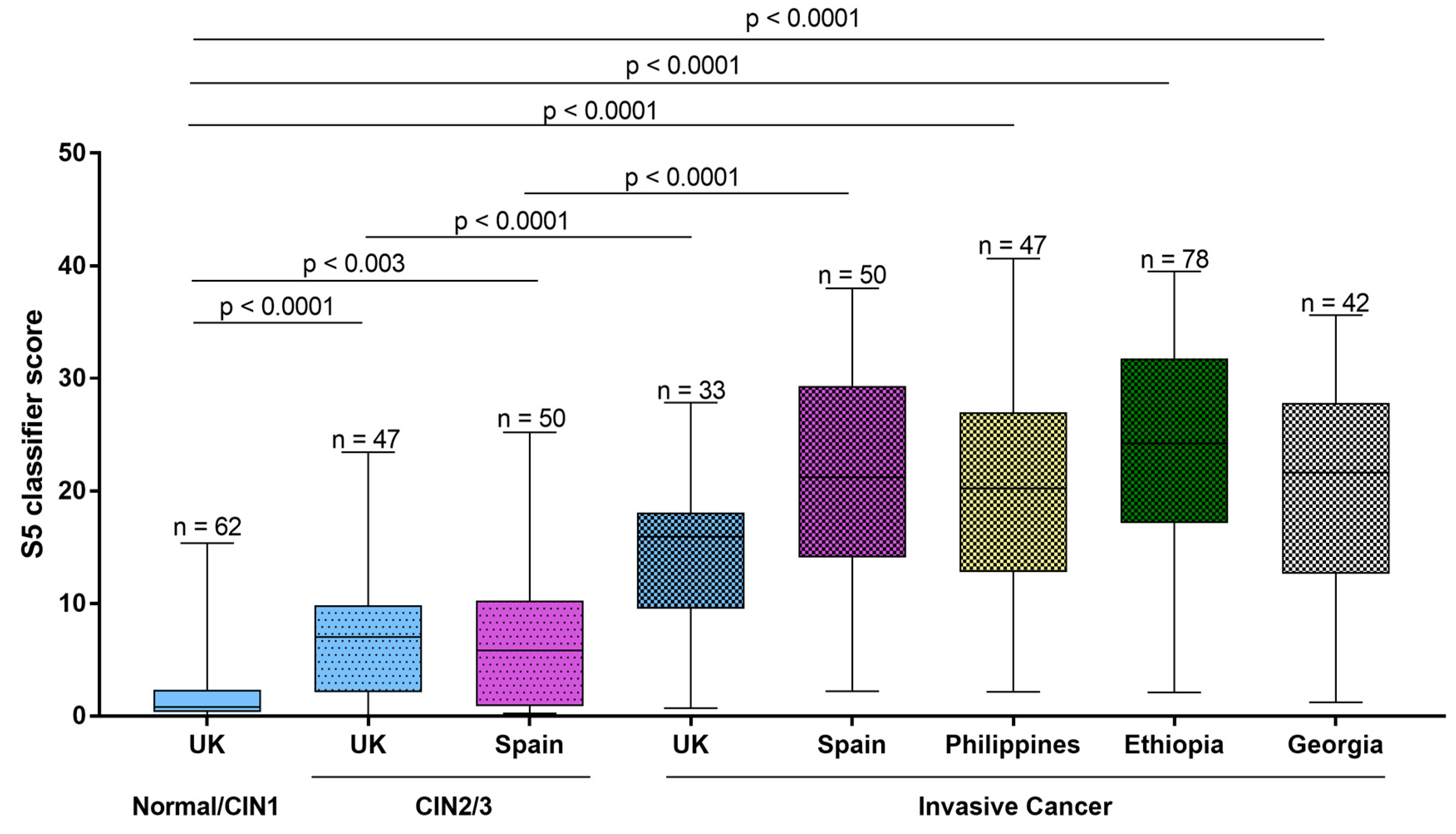
Cervical cancers and CIN2/3 generally have high levels of methylation in selected target human genes and in the HPV genomic late regions,
Figure 1, [
19,
20,
21,
22]. In contrast normal and healthy women with no intraepithelial lesions or malignancy (NILM) usually have very low levels of methylation in these genes [
20]. There is an interesting and useful gradient of DNA methylation that increases over time in women as they progress toward precancer and cancer. For example, a study in Costa Rica showed that temporal elevations in DNA methylation in the HPV16 L1 region predicted development of invasive cancer more than five years in advance [
19]. Recent studies with the S5 DNA methylation test have confirmed these results,
Figure 2 [
23] and shown that invasive cervical cancers associated with HPV16 and HPV18 can be predicted with high sensitivity (8 of 8 cases) up to five years in advance by stepwise increases in DNA methylation values, mostly in HPV L1 DNA regions. Another important DNA methylation assay based on the human target genes FAM19A4 and miR124-2 can also predict cervical cancers in advance. Among 35 cancers diagnosed in women who were hrHPV positive at baseline in the POBASCAM trial, FAM19A4/mir124-2 methylation identified 12 of 12 cancers diagnosed within 4 years of baseline and 12 of 23 cancers within 5 to 14 years of diagnosis [
14,
24].
5. DNA Methylation to Triage Women with hrHPV Infection or Abnormal Cytology
We previously developed a triage classifier called S5, based on pyrosequencing and quantitative measurement of DNA methylation of the late regions of HPV16, HPV18, HPV31, and HPV33, combined with the promoter region of a human gene
EPB41L3 and we demonstrated the good performance of this test in large clinical studies in the USA, Europe, China, and Latin America (
Figure 2 and contained references). A recent meta-analysis provides summarized data on the performance of S5 and other DNA methylation tests. The review concluded that DNA methylation was indeed a good method for detecting cervical cancer and precancers and had promise as an improved new method to triage hrHPV infected women [
25].
6. careREVEAL a New qMSP DNA Methylation Test Evolved from the S5 Classifier
Although S5 is a good research method for DNA methylation testing it is not easy to establish in most laboratories because it requires expensive specialized equipment (the pyrosequencer) and is a relatively labor-intensive assay, taking two days to complete each run. In view of the need for a simpler method we developed a new assay qMSP(EPB/16/18), which has the commercial name careREVEAL (CE) and is a novel qPCR methylation assay intended to triage women with abnormal cytology (primarily ASCUS or LSIL) or women infected with hrHPV. The new assay was trained on large group of precancers (200+) and normal (200+) convenience specimens (data not shown) and was validated in a different screening population of 403 women composed of 45 cases of CIN2 or higher (CIN2+) and 358 controls with NILM/CIN1 (no intraepithelial lesions or malignancy and / or cervical intraepithelial neoplasia grade 1 ― considered low risk lesions). An abbreviated version of the unpublished validation study results is presented in
Table 1. The qMSP(EPB/16/18) assay gave a sensitivity of 90% for CIN2+ and a specificity of 76%, which were both significantly higher by McNemars test than the sensitivity and specificity of either cytology (65% and 67%, respectively) or HPV16/18 genotyping (64% and 67%, respectively).
7. Potential Clinical Algorithms for DNA Methylation Testing
Selected genes in cervical cancer almost always have high or very high levels of methylation, while CIN3 characteristically have somewhat lower DNA methylation. In contrast women with NILM/CIN1 generally have low levels of methylation. These differences in DNA methylation can be used as targets for effective triage to find risky lesions among hrHPV infected women and women with minor cytological abnormalities (
Figure 1 and
Figure 2). As regards use of methylation assays for followup, a small proportion of CIN2 have levels of DNA methylation similar to cancer and CIN3 but most CIN2 have lower levels, comparable to CIN1; a pattern that may be expected from a lesion that seldom progresses to cervical cancer [
26]. In fact, many researchers have questioned whether CIN2 is a distinct lesion entity, as opposed to having an origin as a misclassification of CIN1 or CIN3. Despite these ambiguities in CIN2 it appears that DNA methylation can be used to indicate which CIN2 will progress to CIN3 and cancer versus lesions that will regress or remain indolent [
27]. DNA methylation testing and followup in young women of childbearing age with CIN2 can help to preserve fertility and allow for easier and safer pregnancies.
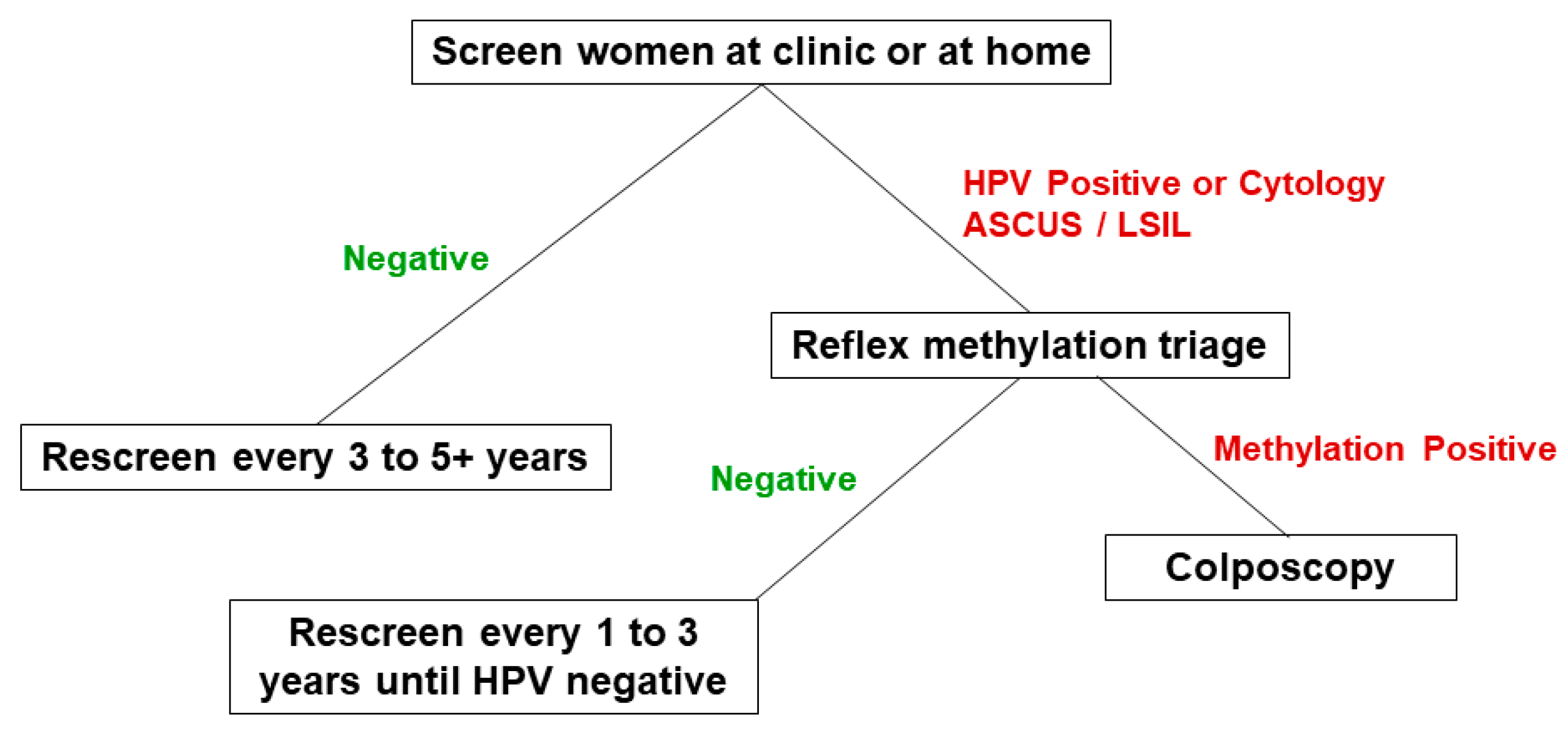
The methylation assay can be run on exfoliated cervical specimens collected by a clinician or self-collected at home, and produces diagnostically useful results regardless of availability of concurrent cytology [
28]. For routine use of the DNA methylation assay we envisage a screening approach that tests for the cocktail of 13 hrHPV types (or uses screening cytology) and then reflex tests for methylation levels of HPVs and
EPB41L3 on hrHPV or cytology positive women. Such a fully integrated molecular screening-triage method would provide the benefit of rapid and more accurate results that can quickly separate women into three management groups: a) negative for all biomarkers, who would go back to routine screening; b) hrHPV (or cytology ASCUS/LSIL) positive and methylation negative, who would have repeat testing, and c) methylation positive regardless of hrHPV (or cytology) status, who would be referred to colposcopy (
Figure 3). In situations where cytology screening is also in use the women with high grade squamous intraepithelial lesions (HSIL) or any other diagnosis indicative or suggestive of precancer or cancer would proceed immediately to colposcopy.
8. Future Directions
Cytology has been slowly giving way to hrHPV DNA screening, with repeat cytology increasingly taking a stronger role as a triage test for hrHPV screen-positive women. HPV16/18 genotyping was incorporated into triage more recently and has provided some benefits, while also sending more women without disease to colpsocopy. Concurrently, prophylactic vaccination is ongoing in developed countries, which is leading to a reduction in the prevalence of certain hrHPV types, especially HPV16 and HPV18, as well as a reduction in the prevalence of precancer and invasive cancer. A consequence of these effects is that other hrHPV types are becoming relatively more important as etiological agents of the remaining cancers. These changes are expected to reduce the value of HPV16/18 genotyping and repeat cytology over time, in other words these triage tests are expected to become less specific. It has long been recognized that a major limitation of HPV testing is mediocre specificity, thus there is a growing need for novel approaches to screening and triage. DNA methylation testing is an important new approach that can provide both enhanced sensitivity and specificity for CIN2/3 and cancer and is a test ready to take on roles in both the triage of screen positive women as well as followup of women with CIN2.
There are several relatively recent commercially available DNA methylation tests that rely on the accuracy, reliability, simplicity, and user familiarity of qPCR technology, among these are the QIAsure, GynTect and careREVEAL tests. These methylation assays have costs similar to other routine PCR tests such as hrHPV screening assays. Methylation assays can be readily incorporated into most diagnostic laboratory settings. Algorithms using DNA methylation on self-collected vaginal specimens and urine specimens collected at home may be suitable for developed countries as well as for some low-resource settings [
19,
28,
30,
31]. These kinds of samples can be stabilized to allow their shipment, which may take days or weeks, and then be tested in central locations. Test positive women would be called into the treatment clinics or alternatively, clinicians could be sent to the places where the women reside, to render suitable treatments.
There are other newer DNA methylation tests in the wings, which show promise for the future. Some such tests will rely on next generation sequencing and allow the interrogations of numerous CpG sites on human gene panels and on all hrHPV types. An interesting candidate approach is the Oxford Nanopore system (Oxford Nanopore Technologies PLC, Oxford UK) which can run assays on native DNA and give quantitative methylation results without the need for the bisulfite conversion step. These tests are still in development and their requirement for large amounts of DNA and their high costs will need to be addressed before they can become useful diagnostic tools in routine clinical laboratories.
Author Contributions
Study conception and design: ATL, XJZ. Hospital and clinic procedures: QYW, RJL. Performance of diagnostics tests and collection of data: XJZ, QYW, RJL, ATL. Statistical analyses: ATL. Literature review: ATL. Drafting of the paper: ATL. Editing of the paper: all authors. Final editing of the paper: ATL, XJZ. Final responsibility for accuracy and details of the paper: ATL.
Funding
This work was supported in part by a grant from Cancer Research UK (Program Grant C569/A10404), by funding from careLYFE, and by donation of time from the staff of Jiangyin Hospital. Funders had no role in the literature review, analysis and interpretation of data, or first drafting of this review. ATL holds final responsibility for all contents in the paper.
Acknowledgments
We gratefully recognize the patients who participated in our study. Special thanks to the staff at Jiangyin Hospital and at careLYFE for assistance with clinical and laboratory procedures. Staff of the Molecular Epidemiology Laboratory at Queen Mary University of London helped to develop the S5 test and worked diligently on the published S5 studies. Many thanks to the head of the institute Jack Cuzick for his unwavering support of ATL’s research at QMUL over the years. Special consideration is due to Cristiana Banila for her strong dedication and excellent laboratory skills in executing the worldwide study of methylation in cervical cancer (Figure 1); Gary Clifford at IARC was also a key participant in this study [
22].
Conflicts of Interest
ATL received travel costs and sponsorships from careLYFE for speaking engagements. XJZ is an employee of careLYFE. Other authors have no conflicts of interest.
References
- Burnett, S.T.; Sleeman, J. Uneven distribution of methylation sites within the human papillomavirus 1a genome: possible relevance to viral gene expression. Nucleic Acids Res. 1984, 12, 8847–8860. [Google Scholar] [CrossRef] [PubMed]
- Rösl, F.; Dürst, M.; zur Hausen, H. Selective suppression of human papillomavirus transcription in non-tumorigenic cells by 5-azacytidine. The EMBO Journal 1988, 7, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. U S A 1983, 80, 3812–3815. [Google Scholar] [CrossRef]
- Boshart, M.; Gissmann, L.; Ikenberg, H.; Kleinheinz, A.; Scheurlen, W.; zur Hausen, H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. The EMBO Journal 1984, 3, 1151–1157. [Google Scholar] [CrossRef]
- Lorincz, A.T.; Lancaster, W.D.; Kurman, R.J.; Jenson, A.B.; Temple, G.F. Characterization of human papillomaviruses in cervical neoplasias and their detection in routine clinical screening. In: Viral Etiology of Cervical Cancer. Banbury Report. Peto, R.; zur Hausen, H. (eds). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory 1986, 225-237.
- Lorincz, A.T.; Lancaster, W.D.; Temple, G.F. Cloning and characterization of the DNA of a new human papillomavirus from a woman with dysplasia of the uterine cervix. J. Virology 1986, 58, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, A.T.; Temple, G.F.; Kurman, R.J.; Jenson, A.B.; Lancaster, W.D. Oncogenic association of specific human papillomavirus types with cervical neoplasia. J. National Cancer Institute 1987, 79, 671–677. [Google Scholar]
- Lorincz, A.T.; Reid, R.; Jenson, A.B.; Greenberg, M.D.; Lancaster, W.; Kurman, R.J. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstetrics and Gynecology 1992, 79, 328–337. [Google Scholar] [CrossRef]
- Bosch, F.X.; Lorincz, A.; Muñoz, N.; Meijer, C.; Shah, K.V. The causal relation between human papillomavirus and cervical cancer. J. Clinical Pathology 2002, 55, 244–265. [Google Scholar] [CrossRef]
- Schiffman, M.; Herrero, R.; Hildesheim, A.; Sherman, M.E.; Bratti, M.; Wacholder, S.; Alfaro, M.; Hutchinson, M.; Morales, J.; Greenberg, M.D.; Lorincz, A.T. HPV DNA testing in cervical cancer screening: results from women in a high-risk province of Costa Rica. JAMA 2000, 283, 87–93. [Google Scholar] [CrossRef]
- Wright, T.C., Jr.; Denny, L.; Kuhn, L.; Pollack, A.; Lorincz, A. HPV DNA testing of self-collected vaginal samples compared with cytologic screening to detect cervical cancer. JAMA 2000, 283, 81–86. [Google Scholar] [CrossRef]
- Khan, M.J.; Castle, P.E.; Lorincz, A.T.; Wacholder, S.; Sherman, M.; Scott, D.R.; Rush, B.B.; Glass, A.G.; Schiffman, M. The elevated 10-Year Risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. National Cancer Institute 2005, 97, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Wright, T.C., Jr.; Massad, L.S.; Dunton, C.J.; Spitzer, M.; Wilkinson, E.J.; Solomon, D. 2006 consensus guidelines for the management of women with abnormal cervical screening tests. J. Low. Genit. Tract. Dis. 2007, 11, 201–222. [Google Scholar] [CrossRef]
- Rijkaart, D.C.; Berkhof, J.; Rozendaal, L.; van Kemenade, F.J.; Bulkmans, N.W.; Heideman, D.A.; Kenter, G.G.; Cuzick, J.; Snijders, P.J.; Meijer, C.J. Human papillomavirus testing for the detection of high-grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2011, 13, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Egemen, D.; Cheung, L.C.; Chen, X.; Demarco, M.; Perkins, R.B.; Kinney, W.; Poitras, N.; Befano, B.; Locke, A.; Guido, R.S.; Wiser, A.L.; Gage, J.C.; Katki, H.A.; Wentzensen, N.; Castle, P.E.; Schiffman, M.; Lorey, T.S. Risk Estimates Supporting the 2019 ASCCP Risk-Based Management Consensus Guidelines. J. Low. Genit. Tract Dis. 2020, 24, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, A.; Castanon, A.; Lim, A.W.W.; Sasieni, P. New strategies for human papillomavirus-based cervical screening. Women’s Health. 2013, 9, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Bergeron, C.; von Knebel Doeberitz, M.; Gravitt, P.; Jeronimo, J.; Lorincz, A.T.; Meijer, C.J.L.; Sankaranarayanan, R.; Snijders, P.J.F.; Szarewski, A. New technologies and procedures for cervical cancer screening. Vaccine 2012, 30, F107–F116. [Google Scholar] [CrossRef] [PubMed]
- Dillner, J.; Rebolj, M.; Birembaut, P.; Petry, K.U.; Szarewski, A.; Munk, C.; de Sanjose, S.; Naucler, P.; Lloveras, B.; Kjaer, S.; Cuzick, J.; van Ballegooijen, M.; et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ 2008, 337, a1754. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Schiffman, M.; Ghosh, A.; Rodriguez, A.C.; Vasiljevic, N.; Wentzensen, N.; Herrero, R.; Hildesheim, A.; Wacholder, S.; Scibior-Bentkowska, D.; Burk, R.D.; Lorincz, A.T. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. International J. Cancer, 2013; 132, 1412–1422. [Google Scholar]
- Lorincz, A.T. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytologica 2016, 60, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, A.T.; Brentnall, A.R.; Scibior-Bentkowska, D.; Reuter, C.; Banwait, R.; Cadman, L.; Austin, J.; Cuzick, J.; Vasiljević, N. Validation of a DNA methylation HPV triage classifier in a screening sample. International J. Cancer 2016, 138, 2745–2751. [Google Scholar] [CrossRef]
- Banila, C.; Lorincz, A.T.; Scibior-Bentkowska, D.; Clifford, G.M.; Kumbi, B.; Beyene, D.; Wheeler, C.M.; Cuschieri, K.; Cuzick, J.; Nedjai, B. Clinical performance of methylation as a biomarker for cervical carcinoma in situ and cancer diagnosis: a worldwide study. International J. Cancer 2022, 150, 290–302. [Google Scholar] [CrossRef]
- Cook, D.A.; Krajden, M.; Brentnall, A.R.; Gondara, L.; Chan, T.; Law, J.H.; Smith, L.W.; van Niekerk, D.J.; Ogilvie, G.S.; Coldman, A.J.; Warman, R.; Reuter, C.; Cuzick, J.; Lorincz, A.T. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. International J. Cancer 2019, 144, 2587–2595. [Google Scholar] [CrossRef]
- De Strooper LMA, Berkhof J, Steenbergen RDM, Lissenberg-Witte BI, Snijders PJF, Meijer C, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124-2 methylation test: A post hoc analysis in the POBASCAM trial with 14 year follow-up. Int J Cancer. 2018 Sep 15;143(6):1541-8.
- Kelly, H.; Benavente, Y.; Pavon, M.A.; De Sanjose, S.; Mayaud, P.; Lorincz, A.T. Performance of DNA methylation assays for detection of high-grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta-analysis. British J. Cancer 2019, 121, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Nasiell, K.; Nasiell, M.; Vaclavinkova, V. Behavior of moderate cervical dysplasia during long-term follow-up. Obstetrics and Gynecology 1983, 61, 609–614. [Google Scholar] [PubMed]
- Louvanto, K.; Aro, K.; Nedjai, B.; Bützow, R.; Jakobsson, M.; Kalliala, I.; Dillner, J.; Nieminen, P.; Lorincz, A. Methylation in predicting progression of untreated high-grade cervical intraepithelial neoplasia. Clinical Infectious Diseases 2020, 70, 2582–2590. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, V.M.; Bosgraaf, R.P.; van Kemenade, F.J.; Rozendaal, L.; Heideman, D.A.; Hesselink, A.T.; Bekkers, R.L.; Steenbergen, R.D.; Massuger, L.F.; Melchers, W.J.; Bulten, J.; Overbeek, L.I. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): a randomised controlled non-inferiority trial. Lancet Oncol. 2014, 15, 315–322. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, X.; Hu, S.; Chen, S.; Zhao, S.; Dong, L.; Carvalho, A.L.; Muwonge, R.; Zhao, F.; Basu, P. Triage performance and predictive value of the human gene methylation panel among women positive on self-collected HPV test: Results from a prospective cohort study. International J. Cancer 2022, 151, 878–887. [Google Scholar] [CrossRef]
- Lazcano-Ponce, E.; Lorincz, A.T.; Cruz-Valdes, A.; Salmeron, J.; Uribe, P.; Velasco-Mondragón, E.; Hernandez Nevarez, P.; Diaz Acosta, R.; Hernández-Avila, M. Self-collection of vaginal specimens for human papillomavirus testing in cervical cancer prevention (MARCH): a community-based randomised controlled trial. Lancet 2011, 378, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Serrano, B.; Ibanez, R.; Robles, C.; Peremiquel-Trillas, P.; de Sanjose, S.; Bruni, L. Worldwide use of HPV self-sampling for cervical cancer screening. Prev. Med. 2022, 154, 106900. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, A.T.; Sánchez, G.I.; Nedjai, B.; Agudelo, M.C.; Brentnall, A.R.; Cuschieri, K.; Castañeda, K.M.; Cuzick, J.; Lorincz, A.T.; ASC-US-COL Trial Group. Effective methylation triage of HPV positive women with abnormal cytology in a middle-income country. International J. Cancer 2021, 148, 1383–1393. [Google Scholar] [CrossRef]
- Kelly, H.A.; Chikandiwa, A.; Warman, R.; Segondy, M.; Sawadogo, B.; Vasiljevic, N.; Didelot, M.N.; Meda, N.; Weiss, H.A.; Delany-Moretlwe, S.; Mayaud, P.; Lorincz, A. Associations of human gene EPB41L3 DNA methylation and cervical intraepithelial neoplasia in women living with HIV-1 in Africa. Aids. 2018, 32, 2227–2236. [Google Scholar] [CrossRef]
- Lorincz, A.T.; Brentnall, A.R.; Vasiljević, N.; Scibior-Bentkowska, D.; Castanon, A.; Fiander, A.; Powell, N.; Tristram, A.; Cuzick, J.; Sasieni, P. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. International J. Cancer 2013, 133, 637–644. [Google Scholar] [CrossRef]
- Brentnall, A.R.; Vasiljevic, N.; Scibior-Bentkowska, D.; Cadman, L.; Austin, J.; Cuzick, J.; Lorincz, A.T. HPV33 DNA methylation measurement improves cervical pre-cancer risk estimation of an HPV16, HPV18, HPV31 and EPB41L3 methylation classifier. Cancer Biomarkers 2015, 15, 669–675. [Google Scholar] [CrossRef]
- Brentnall, A.R.; Vasiljević, N.; Scibior-Bentkowska, D.; Cadman, L.; Austin, J.; Szarewski, A.; Cuzick, J.; Lorincz, A.T. A DNA methylation classifier of cervical precancer based on human papillomavirus and human genes. International J. Cancer 2014, 135, 1425–1432. [Google Scholar] [CrossRef]
- Louvanto, K.; Franco, E.L.; Ramanakumar, A.V.; Vasiljević, N.; Scibior-Bentkowska, D.; Koushik, A.; Cuzick, J.; Coutlée, F.; Lorincz, A.T.; Biomarkers of Cervical Cancer Risk Study Team. Methylation of viral and host genes and severity of cervical lesions associated with human papillomavirus type 16. International J. Cancer 2015, 136, E638–E645. [Google Scholar] [CrossRef]
- Vasiljevic, N.; Scibior-Bentkowska, D.; Brentnall, A.; Cuzick, J.; Lorincz, A. A comparison of methylation levels in HPV18, HPV31 and HPV33 genomes reveals similar associations with cervical precancers. J. Clinical Virology 2014, 59, 161–166. [Google Scholar] [CrossRef]
- Vasiljevic, N.; Scibior-Bentkowska, D.; Brentnall, A.R.; Cuzick, J.; Lorincz, A. Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high-risk HPV positive women. Gynecology and Oncology 2014, 132, 709–714. [Google Scholar] [CrossRef]
- Wentzensen, N.; Sun, C.; Ghosh, A.; Kinney, W.; Mirabello, L.; Wacholder, S.; Shaber, R.; LaMere, B.; Clarke, M.; Lorincz, A.T.; Castle, P.E.; Schiffman, M.; Burk, R.D. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. J. National Cancer Institute 2012, 104, 1738–1749. [Google Scholar] [CrossRef]
- Gu, Y.Y.; Zhou, G.N.; Wang, Q.; Ding, J.X.; Hua, K.Q. Evaluation of a methylation classifier for predicting pre-cancer lesion among women with abnormal results between HPV16/18 and cytology. Clinical Epigenetics 2020, 12, 57. [Google Scholar] [CrossRef]
- Adcock, R.; Nedjai, B.; Lorincz, A.T.; Scibior-Bentkowska, D.; Banwait, R.; Torrez-Martinez, N.; Robertson, M.; Cuzick, J.; Wheeler, C.M.; New Mexico HPV Pap Registry Steering Committee. DNA methylation testing with S5 for triage of high-risk HPV positive women. International J. Cancer 2022, 151, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Hernández-López, R.; Lorincz, A.T.; Torres-Ibarra, L.; Reuter, C.; Scibior-Bentkowska, D.; Warman, R.; Nedjai, B.; Mendiola-Pastrana, I.; León-Maldonado, L.; Rivera-Paredez, B.; Ramírez-Palacios, P.; Lazcano-Ponce, E.; Cuzick, J.; Salmerón, J. Methylation estimates the risk of precancer in HPV-infected women with discrepant results between cytology and HPV16/18 genotyping. Clinical Epigenetics 2019, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.R.; Nedjai, B.; Lorincz, A.T.; Schell, M.J.; Rahman, S.; Rawinder, B.; Boulware, D.; Sirak, B.; Martin-Gomez, L.; Abrahamsen, M.; Isaacs-Soriano, K.A.; Wenig, B.; Chung, C.H.; Caudell, J. Methylation of HPV 16 and EPB41L3 in oral gargles: Associations with oropharyngeal cancer detection and tumor characteristics. International J. Cancer 2020, 146, 1018–1030. [Google Scholar] [CrossRef]
- Lorincz, A.T.; Nathan, M.; Reuter, C.; Warman, R.; Thaha, M.A.; Sheaff, M.; Vasiljevic, N.; Ahmad, A.; Cuzick, J.; Sasieni, P. Methylation of HPV and a tumor suppressor gene reveals anal cancer and precursor lesions. Oncotarget 2017, 8, 50510. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).