Submitted:
25 October 2023
Posted:
26 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and discussion
2.1. 1H NMR
2.2. Molecular modeling
2.2.1. Atomic charges, bond lengths and bond angles
2.3. Characterization of metal complexes
2.4. IR spectral studies.
2.5. Molar conductivity measurements
2.6. Electronic spectra and magnetic moment measurements
2.7. Mass spectral study
2.8. Thermogravimetric analysis data
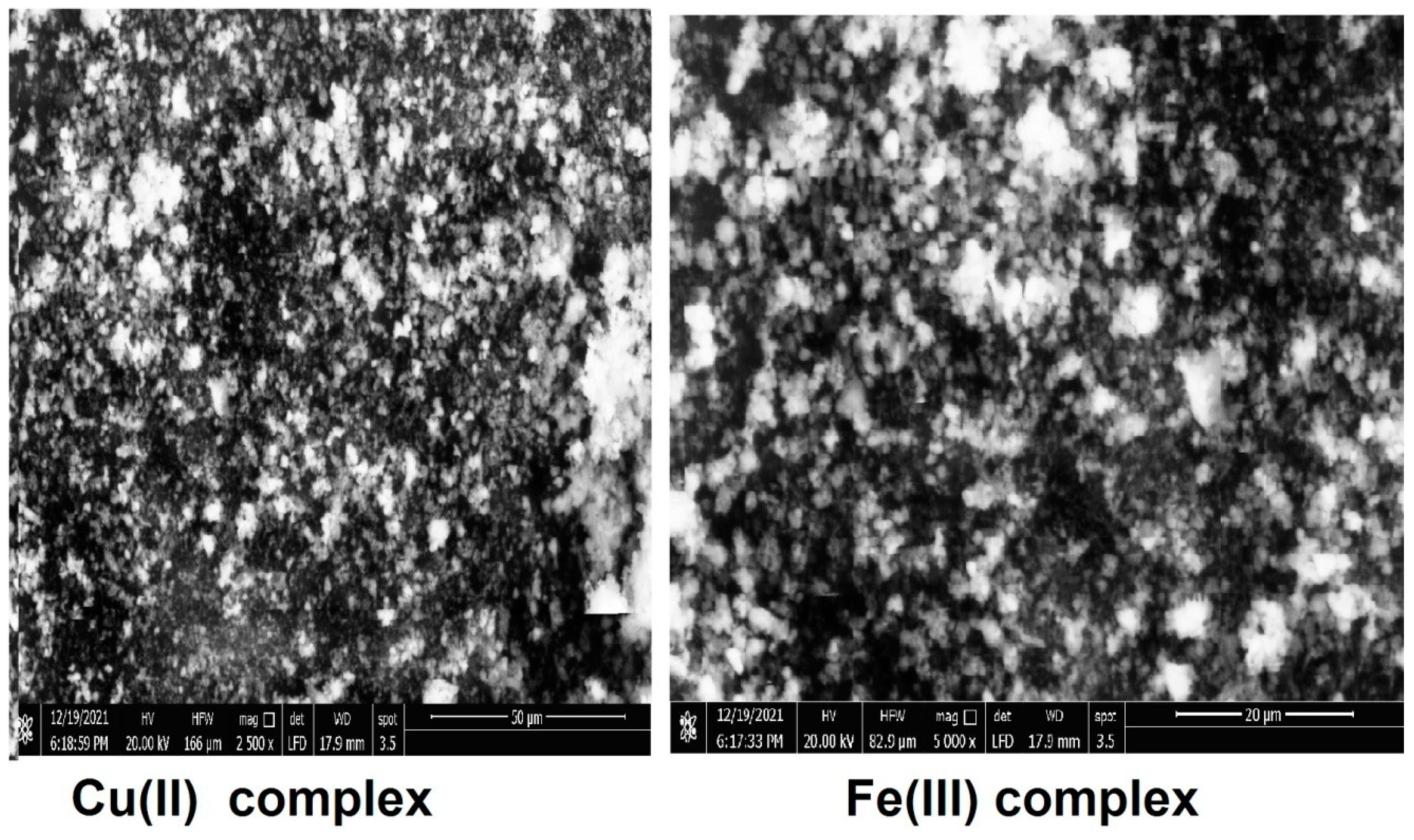
2.9. SEM analysis
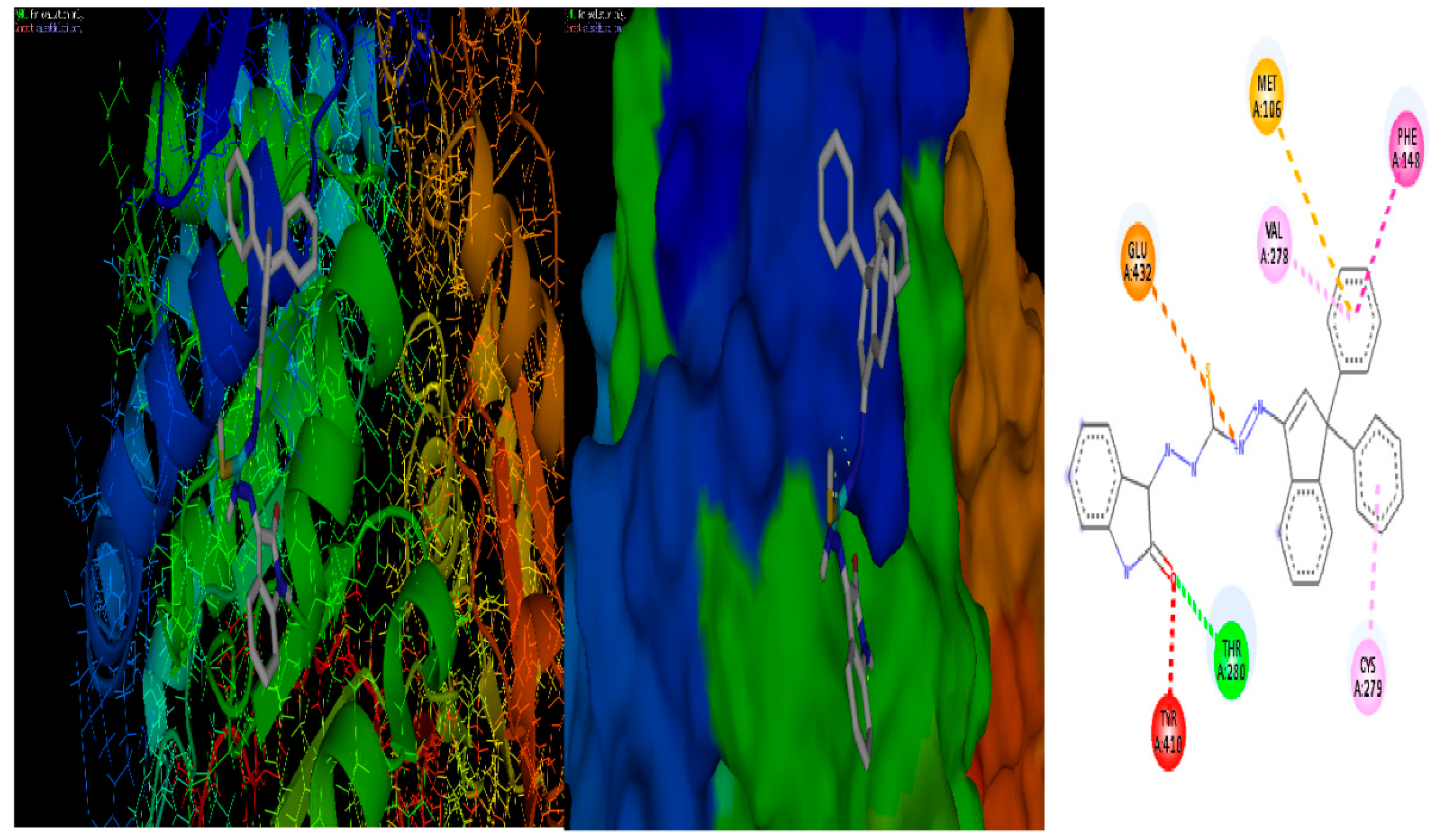
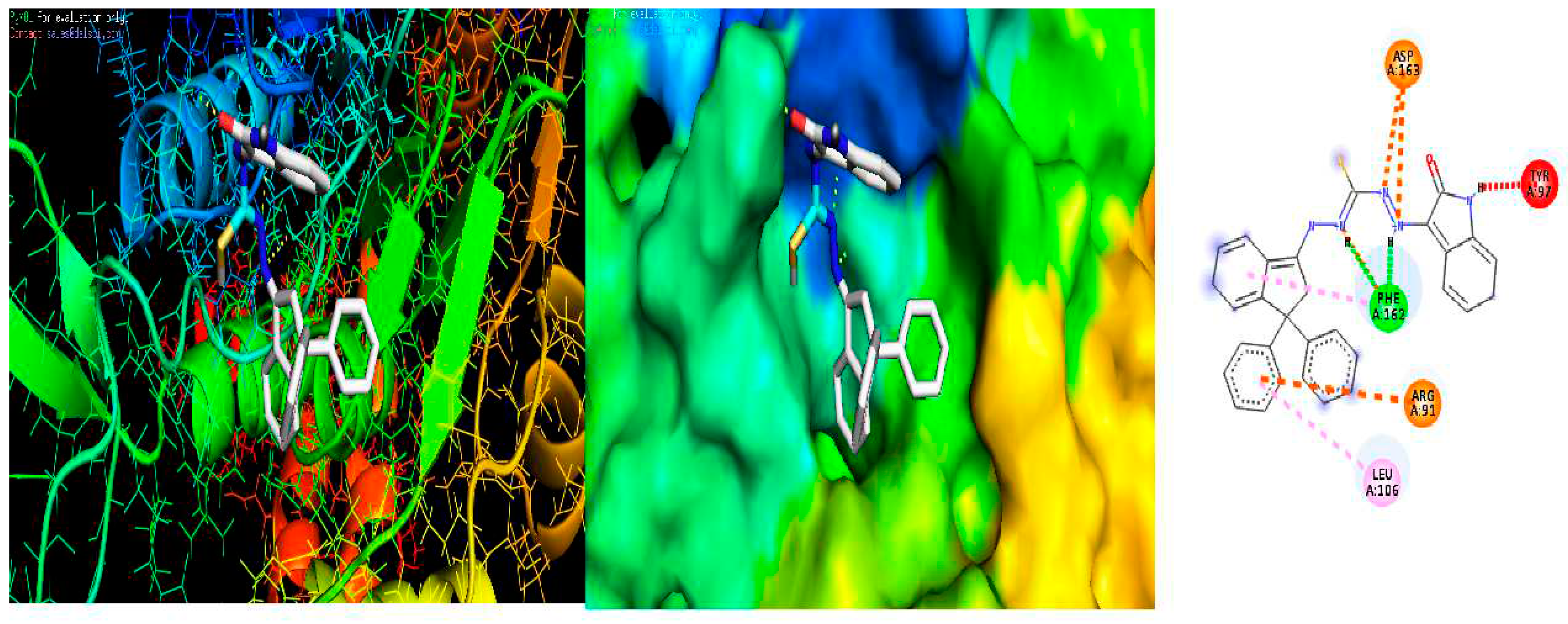
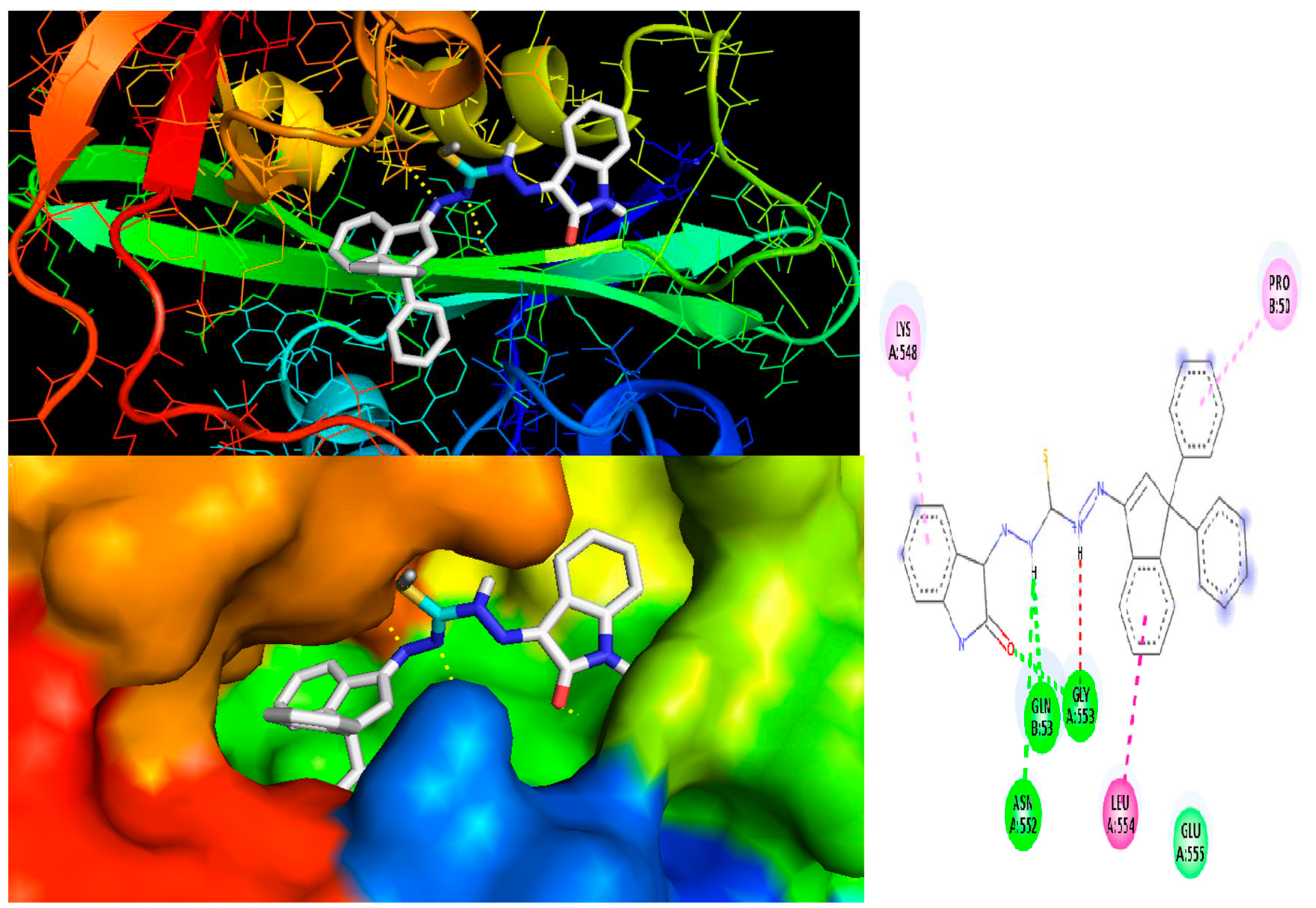
2.10. Molecular Docking
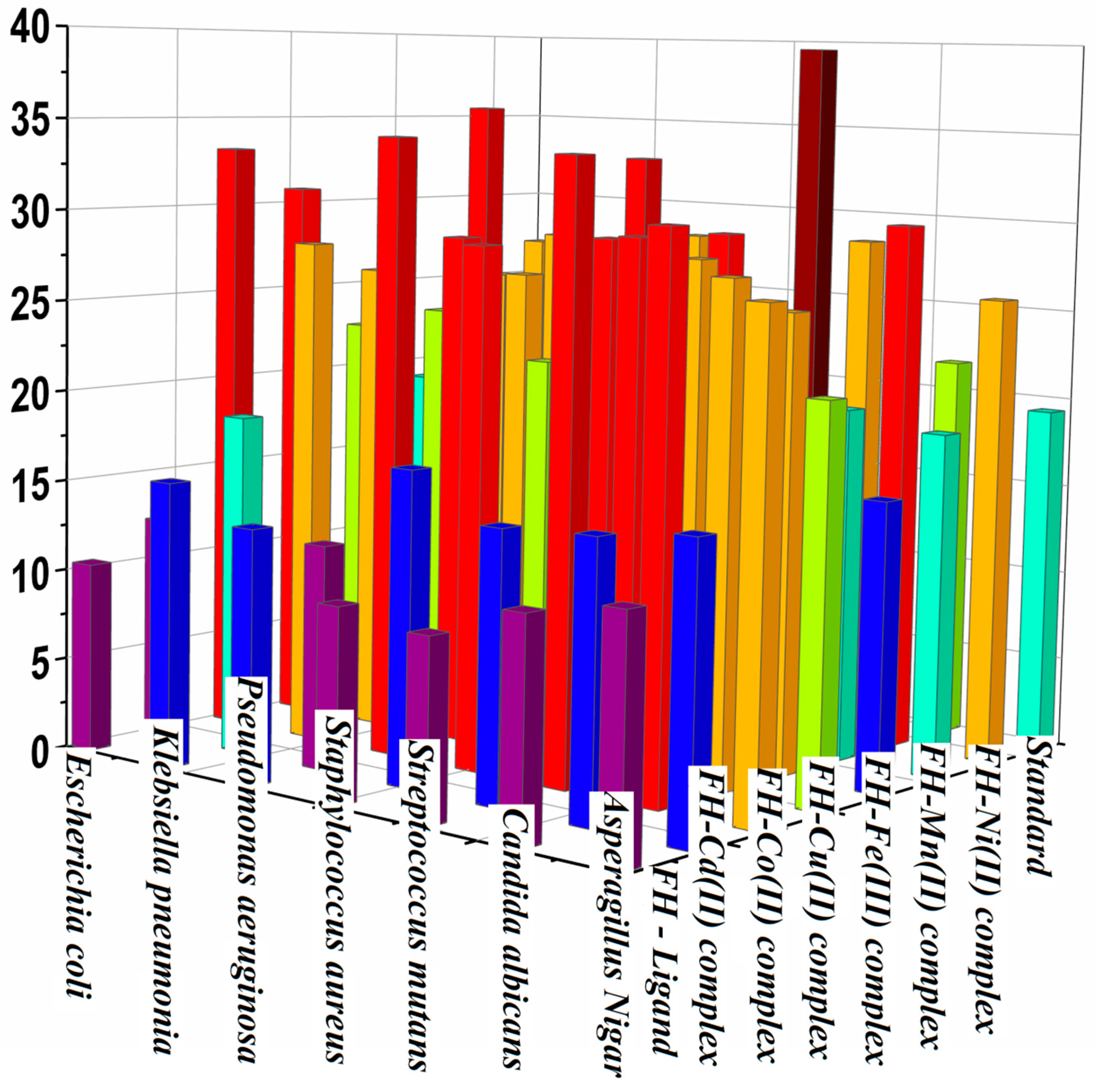
2.11. Antimicrobial activity of hydrazones ligand and its metal complexes
3. Experimental
3.1. Materials and reagents
3.2. Solutions
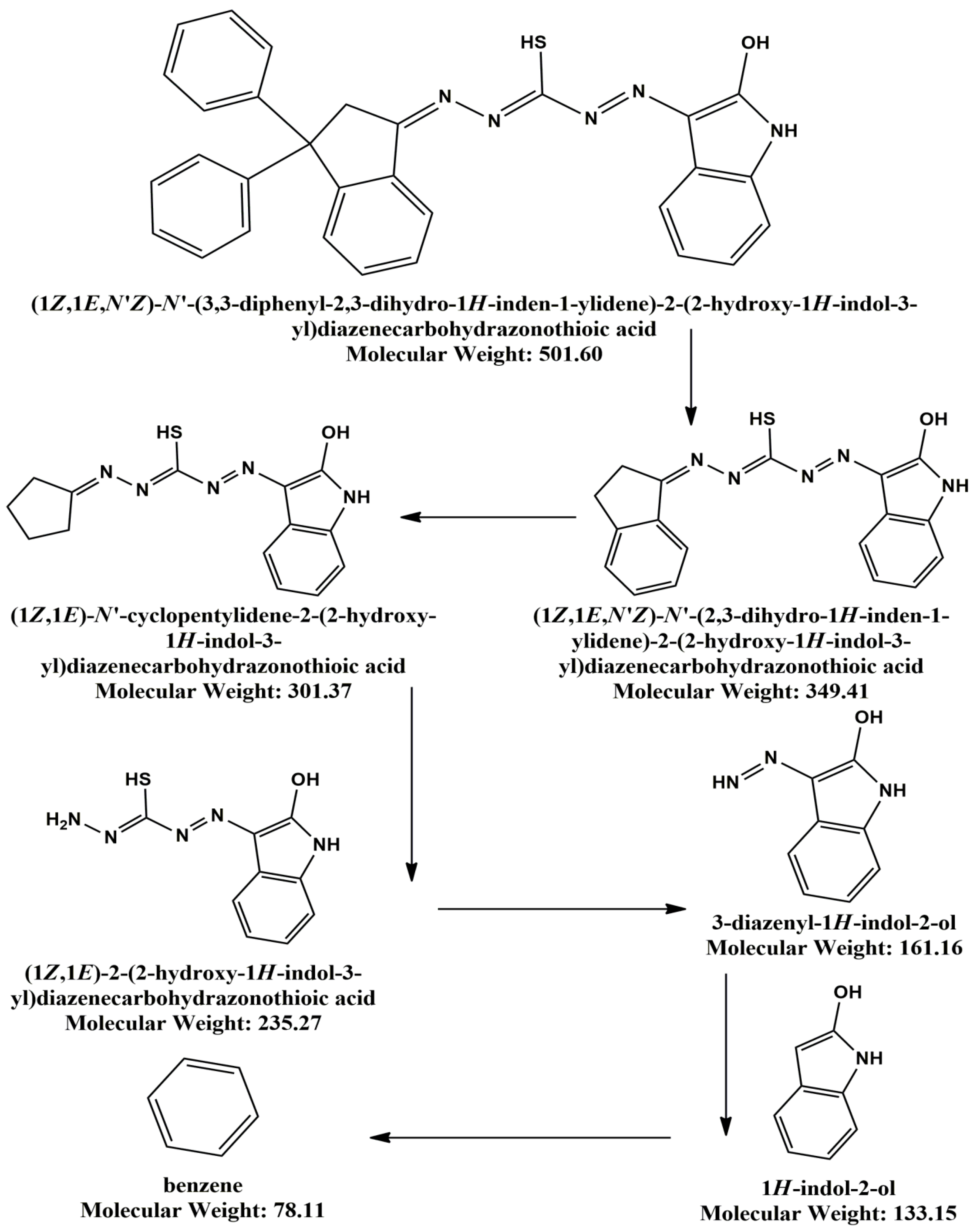
3.3. Synthesis of new Schiff bases based on 3, 3-diphenyl-2,3-dihydro-1H-inden-1-one and thiocarbohydrazone derivatives ligand
3.3.1. Synthesis of 3, 3-diphenyl-2, 3-dihydro-1H-inden-1-one - β -thiocarbohydrazone 3:
3.3.2. N′-((E)-3,3-diphenyl-2,3-dihydro-1H-inden-1-ylidene)-2-((Z)-2-oxoindolin-3-ylidene)hydrazine-1-carbothiohydrazide 5:
3.4. Synthesis of metal complexes
3.5. Spectrophotometric studies
3.6. Molecular docking
3.7. Biological activity
3.8. Computational methodology
4. Conclusion
Supplementary Materials
References
- Muhammad, M.T.; Ghouri, N.; Khan, K.M.; Choudhary, M.I.; Perveen, S. Synthesis of Thiocarbohydrazones and Evaluation of their in vitro Antileishmanial Activity. Med. Chem. 2018, 14, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, A.Y.; Ammar, Y.A.; Abu-Elghait, M.; Salem, M.A.; Assiri, M.A.; Ali, T.E.; et al. Development of novel indolin-2-one derivative incorporating thiazole moiety as DHFR and quorum sensing inhibitors: Synthesis, antimicrobial, and antibiofilm activities with molecular modelling study. Bioorg. Chem. 2022, 119, 105571. [Google Scholar] [CrossRef] [PubMed]
- Ragab, A.; Ammar, Y.A.; Ezzat, A.; Mahmoud, A.M.; Mohamed, M.B.I.; Abdou, S.; et al. Synthesis, characterization, thermal properties, antimicrobial evaluation, ADMET study, and molecular docking simulation of new mono Cu (II) and Zn (II) complexes with 2-oxoindole derivatives. Comput. Biol. Med. 2022, 145, 105473. [Google Scholar] [CrossRef] [PubMed]
- Divya, K.; Pinto, G.M.; Pinto, A.F. Application of metal complexes of Schiff bases as an antimicrobial drug: a review of recent works. Int. J. Curr. Pharm. Res. 2017, 9, 27–30. [Google Scholar] [CrossRef]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. MedChemComm 2018, 9, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, M.C.S.; Fabiani, M.; Castro, E.F.; Cavallaro, L.V.; Finkielsztein, L.M. Synthesis, antiviral evaluation and molecular docking studies of N4-aryl substituted/unsubstituted thiosemicarbazones derived from 1-indanones as potent anti-bovine viral diarrhea virus agents. Bioorg. Med. Chem. 2017, 25, 4055–4063. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.S.M.; Wei, O.C.; Fei, Y.M. Synthesis, characterisation and vasolidation properties of indanone-based chalcones. J. Phys. Sci. 2018, 29, 99–106. [Google Scholar] [CrossRef]
- Medvedev, A.; Buneeva, O.; Kopylov, A.; Tikhonova, O.; Medvedeva, M.; Nerobkova, L.; et al. Brain mitochondrial subproteome of Rpn10-binding proteins and its changes induced by the neurotoxin MPTP and the neuroprotector isatin. Biochem. 2017, 82, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Guo, H. Isatin derivatives and their anti-bacterial activities. Eur. J. Med. Chem. 2019, 164, 678–688. [Google Scholar] [CrossRef]
- Pradeep, S.D.; Gopalakrishnan, A.K.; Manoharan, D.K.; Soumya, R.S.; Gopalan, R.K.; Mohanan, P.V. Isatin derived novel Schiff bases: An efficient pharmacophore for versatile biological applications. J. Mol. Struct. 2023, 1271, 134121. [Google Scholar] [CrossRef]
- Zayed, E.M.; Mohamed, G.G.; Hindy, A.M. Transition metal complexes of novel Schiff base: Synthesis, spectroscopic characterization, and in vitro antimicrobial activity of complexes. J. Therm. Anal. Calorim. 2015, 120, 893–903. [Google Scholar] [CrossRef]
- Ramadan, R.M.; Al-Nasr, A.K.A.; Noureldeen, A.F. Synthesis, spectroscopic studies, antimicrobial activities and antitumor of a new monodentate V-shaped Schiff base and its transition metal complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 417–422. [Google Scholar] [CrossRef]
- Matin, S.; Khojasteh, R. Synthesis, characterization, and antibacterial activities of Cr (III), Co (III), Ni (II), and Mn (III) complexes of heptadentate Schiff base ligand derived from tris (2-aminoethyl) amine. Russ. J. Gen. Chem. 2015, 85, 1763–1767. [Google Scholar] [CrossRef]
- Rajee, A.O.; Babamale, H.F.; Lawal, A.; Aliyu, A.A.; Osunniran, W.A.; Sheriff, A.O.; et al. Mn (II), Co (II), Ni (II), and Cu (II) complexes of amino acid derived Schiff base ligand: Synthesis, characterization and in-vitro antibacterial investigations. Bull. Chem. Soc. Ethiop. 2021, 35, 97–106. [Google Scholar] [CrossRef]
- Abdel-Rhman, M.H.; El-Asmy, A.A.; Ibrahim, R.; Hosny, N.M. New Schiff base ligand and some of its coordination compounds: Synthesis, spectral, molecular modeling and biological studies. J. Mol. Struct. 2023, 1279, 135023. [Google Scholar] [CrossRef]
- Chang, C.; Chen, W.; Chen, Y.; Chen, Y.; Chen, Y.; Ding, F.; et al. Recent progress on two-dimensional materials. Acta Phys. Chim. Sin. 2021, 37, 2108017. [Google Scholar]
- Mahadevi, P.; Sumathi, S.; Metha, A.; Singh, J. Synthesis, spectral, antioxidant, in vitro cytotoxicity activity and thermal analysis of Schiff base metal complexes with 2, 2′-Bipyridine-4, 4′-dicarboxylic acid as co-ligand. J. Mol. Struct. 2022, 1268, 133669. [Google Scholar] [CrossRef]
- Zayed, E.M.; Mohamed, G.G.; Hassan, W.M.; Elkholy, A.K.; Moustafa, H. Spectroscopic, thermal, biological activity, molecular docking and density functional theoretical investigation of novel bis Schiff base complexes. Appl. Organomet. Chem. 2018, 32, e4375. [Google Scholar] [CrossRef]
- Yadav, M.; Sharma, S.; Devi, J. Designing, spectroscopic characterization, biological screening and antioxidant activity of mononuclear transition metal complexes of bidentate Schiff base hydrazones. J. Chem. Sci. 2021, 133, 1–22. [Google Scholar] [CrossRef]
- Radha, V.P.; Chitra, S.; Jonekirubavathi, S.; Chung, I.-M.; Kim, S.-H.; Prabakaran, M. Transition metal complexes of novel binuclear Schiff base derived from 3, 3′-diaminobenzidine: synthesis, characterization, thermal behavior, DFT, antimicrobial and molecular docking studies. J. Coord. Chem. 2020, 73, 1009–1027. [Google Scholar] [CrossRef]
- Farag, A.M.; Sokker, H.H.; Zayed, E.M.; Eldien, F.A.N.; Abd Alrahman, N.M. Removal of hazardous pollutants using bifunctional hydrogel obtained from modified starch by grafting copolymerization. Int. J. Biol. Macromol. 2018, 120, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- Manan, M.A.F.A.; Mohammat, M.F. Synthesis, structural studies and antimicrobial evaluation of nickel (II) bis-complex of Schiff base of S-benzyldithiocarbazate. Trends in Sciences 2022, 19, 1500. [Google Scholar] [CrossRef]
- Pedro, K.C.N.R.; Ferreira, I.E.P.; Henriques, C.A.; Langone, M.A.P. Enzymatic fatty acid ethyl esters synthesis using acid soybean oil and liquid lipase formulation. Chem. Eng. Commun. 2020, 207, 43–55. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, C.; Gao, X.; Yao, Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 2022, 12, 2115. [Google Scholar] [CrossRef] [PubMed]
- Sudha, A. Evaluation of characterization, biological and computational studies of new Schiff base ligand and some metal (II) complexes. Inorganica Chim. Acta 2022, 534, 120817. [Google Scholar] [CrossRef]
- Zayed, E.M.; Zayed, M.A.; Radwan, M.A.; Alminderej, F.M. Synthesis, characterization, antimicrobial, and docking study of novel 1-(furanyl)-3-(pyrrolyl) propenone-based ligand and its chelates of 3d-transition metal ions. Appl. Organomet. Chem. 2022, 36, e6489. [Google Scholar] [CrossRef]
- Eldeken, G.A.; El-Samahy, F.A.; Zayed, E.M.; Osman, F.H.; Elgemeie, G.E. Synthesis, biological activities and molecular docking analysis of a novel series of 11H-indeno [1, 2-b] quinoxalin-11-one derivatives. J. Mol. Struct. 2022, 1261, 132929. [Google Scholar] [CrossRef]
- Zayed, E.M.; Zayed, M.A.; Hindy, A.M.; Mohamed, G.G. Coordination behaviour and biological activity studies involving theoretical docking of bis-Schiff base ligand and some of its transition metal complexes. Appl. Organomet. Chem. 2018, 32, e4603. [Google Scholar] [CrossRef]
- Çelik, F.; Ünver, Y. 1, 2, 4-Triazole derivative containing thiophen ring: Comparison of theoretical IR and NMR data with experimental. J. Indian Chem. Soc. 2022, 99, 100455. [Google Scholar] [CrossRef]
- Zayed, E.M.; El-Samahy, F.A.; Mohamed, G.G. Structural, spectroscopic, molecular docking, thermal and DFT studies on metal complexes of bidentate orthoquinone ligand. Appl. Organomet. Chem. 2019, 33, e5065. [Google Scholar] [CrossRef]
- Ahmed, Y.M.; Mohamed, G.G. New Tin (IV) Schiff base complexes: synthesis, characterization and antibacterial investigation, docking and theoretical studies. Inorg. Chem. Commun. 2022, 144, 109864. [Google Scholar] [CrossRef]
- Zayed, E.M.; Mohamed, G.G.; Abd El Salam, H.A. Ni (II), Co (II), Fe (III), and Zn (II) mixed ligand complexes of indoline-dione and naphthalene-dione: Synthesis, characterization, thermal, antimicrobial, and molecular modeling studies. Inorg. Chem. Commun. 2023, 147, 110276. [Google Scholar] [CrossRef]
- Kumar, S.; Devi, J.; Dubey, A.; Kumar, D.; Jindal, D.K.; Asija, S.; et al. Co (II), Ni (II), Cu (II) and Zn (II) complexes of Schiff base ligands: Synthesis, characterization, DFT, in vitro antimicrobial activity and molecular docking studies. Res. Chem. Intermed. 2023, 49, 939–965. [Google Scholar] [CrossRef]
- Zayed, E.M.; Zayed, M.A.; Abd El Salam, H.A.; Nawwar, G.A. Synthesis, structural characterization, density functional theory (B3LYP) calculations, thermal behaviour, docking and antimicrobial activity of 4-amino-5-(heptadec-8-en-1-yl)-4H-1, 2, 4-triazole-3-thiol and its metal chelates. Appl. Organomet. Chem. 2018, 32, e4535. [Google Scholar] [CrossRef]
- Kanagasabapathy, G.; Britto, S.; Anbazhagan, V. Synthesis, characterization and molecular docking studies of highly functionalized and biologically active derivatives of 2-aminothiazole. J. Mol. Struct. 2023, 1275, 134593. [Google Scholar] [CrossRef]
- Hassaballah, A.I.; El-Ziaty, A.K.; Ewies, E.F.; Zayed, E.M.; Mohamed, G.G. Synthesis of pyrimidine ligand and its mononuclear metal (II)/(III) complexes: Spectroscopic characterization, thermal, DFT, molecular docking, antimicrobial and anticancer studies. Inorg. Chem. Commun. 2023, 155, 110989. [Google Scholar] [CrossRef]
- Ali, A.; Pervaiz, M.; Saeed, Z.; Younas, U.; Bashir, R.; Ullah, S.; et al. Synthesis and biological evaluation of 4-dimethylaminobenzaldehyde derivatives of Schiff bases metal complexes: A review. Inorg. Chem. Commun. 2022, 145, 109903. [Google Scholar] [CrossRef]
- Moustafa, G.; Sabry, E.; Zayed, E.M.; Mohamed, G.G. Structural characterization, spectroscopic studies, and molecular docking studies on metal complexes of new hexadentate cyclic peptide ligand. Appl. Organomet. Chem. 2022, 36, e6515. [Google Scholar] [CrossRef]
- Gopichand, K.; Mahipal, V.; Rao, N.N.; Ganai, A.M.; Rao, P.V. Co (II), Ni (II), Cu (II), and Zn (II) complexes with Benzothiazole Schiff base ligand: Preparation, Spectral Characterization, DNA Binding, and In Vitro Cytotoxic Activities. Results Chem. 2023, 5, 100868. [Google Scholar] [CrossRef]
- Zayed, E.M.; Abd ElSallam, H.A.; Fathala, M.I.; Sroor, F.M. Naphthalene-1, 2-dione and indoline-2, 3-dione as Convenient Ligands to Prepare Mixed Ligand Complexes of Mn, Cu And Cd: Synthesis, Spectroscopic Characterization, DFT Studyand Antibacterial Activity. ChemistrySelect 2023, 8, e202300145. [Google Scholar] [CrossRef]
- Abd El Salam, H.A.; Mohamed, G.G.; Zayed, E.M. Synthesis, spectroscopic characterization, biological application and molecular docking studies of some transition metal complexes of isophthalamide ligand. J. Mol. Struct. 2023, 1273, 134231. [Google Scholar] [CrossRef]
- Abd El Salam, H.A.; Moustafa, G.; Zayed, E.M.; Mohamed, G.G. Isophthaloylbis (Azanediyl) Dipeptide Ligand and Its Complexes: Structural Study, Spectroscopic, Molecular Orbital, Molecular Docking, and Biological Activity Properties. Polycycl. Aromat. Compd. 2022, 1–23. [Google Scholar] [CrossRef]
- Benabid, W.; Ouari, K.; Bendia, S.; Bourzami, R.; Ali, M.A. Crystal structure, spectroscopic studies, DFT calculations, cyclic voltammetry and biological activity of a copper (II) Schiff base complex. J. Mol. Struct. 2020, 1203, 127313. [Google Scholar] [CrossRef]
- Zayed, E.M.; Ewies, E.F.; Hassaballah, A.I.; Mohamed, G.G. Synthesis, characterization, DFT, docking, antimicrobial and thermal study of pyrimidine-carbonitrile ligand and its metal complexes. J. Mol. Struct. 2023, 1284, 135396. [Google Scholar] [CrossRef]
- Zayed, E.M.; Zayed, M. Synthesis of novel Schiff’s bases of highly potential biological activities and their structure investigation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 143, 81–90. [Google Scholar] [CrossRef]
- Abd El Salam, H.A.; Fathy, U.; Zayed, E.M.; El Shehry, M.F.; Ahmed, E. Gouda, a. Design, Synthesis, Cytotoxic Activity and Molecular Docking Studies of Naphthyl Pyrazolyl Thiazole Derivatives as Anticancer Agents. ChemistrySelect 2023, 8, e202203956. [Google Scholar] [CrossRef]
- Zayed, E.M.; Zayed, M.; El-Desawy, M. Preparation and structure investigation of novel Schiff bases using spectroscopic, thermal analyses and molecular orbital calculations and studying their biological activities. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2015, 134, 155–164. [Google Scholar] [CrossRef]
- Reiss, A.; Cioateră, N.; Dăbuleanu, I.; Chifiriuc, M.C.; Amzoiu, E.; Rotaru, P. New metal (II) complexes with ceftazidime Schiff base. J. Therm. Anal. Calorim. 2018, 131. [Google Scholar] [CrossRef]
- Olar, R.; Badea, M.; Ferbinteanu, M.; Stanica, N.; Alan, I. Spectral, magnetic and thermal characterization of new Ni (II), Cu (II), Zn (II) and Cd (II) complexes with a bischelate Schiff base. J. Therm. Anal. Calorim. 2017, 127, 709–719. [Google Scholar] [CrossRef]
- Zarafu, I.; Olar, R.; Chifiriuc, M.C.; Bleotu, C.; Ioniţă, P.; Mulţescu, M.; et al. Synthesis, thermal, spectral, antimicrobial and cytotoxicity profile of the Schiff bases bearing pyrazolone moiety and their Cu (II) complexes. J. Therm. Anal. Calorim. 2018, 134, 1851–1861. [Google Scholar] [CrossRef]
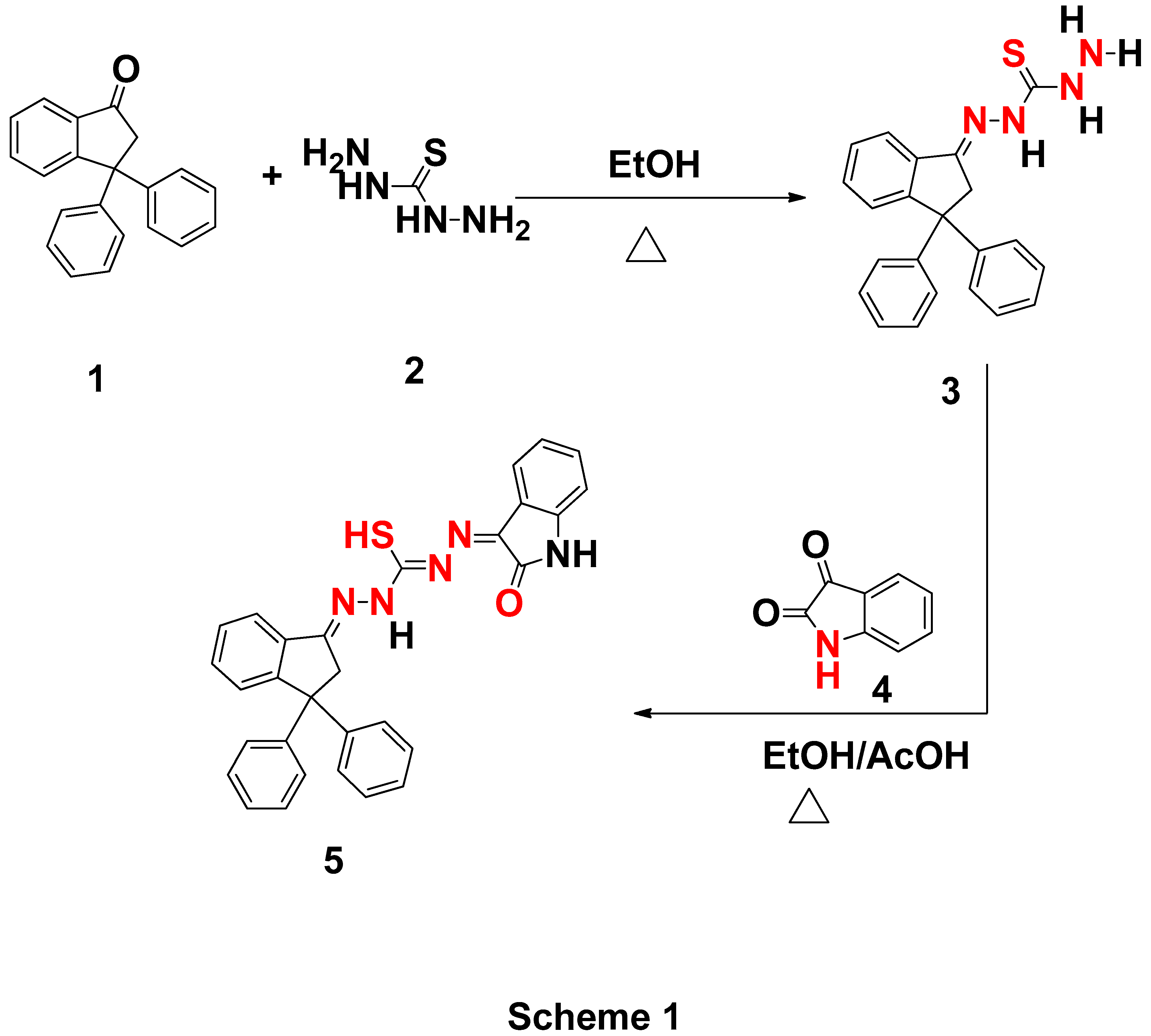
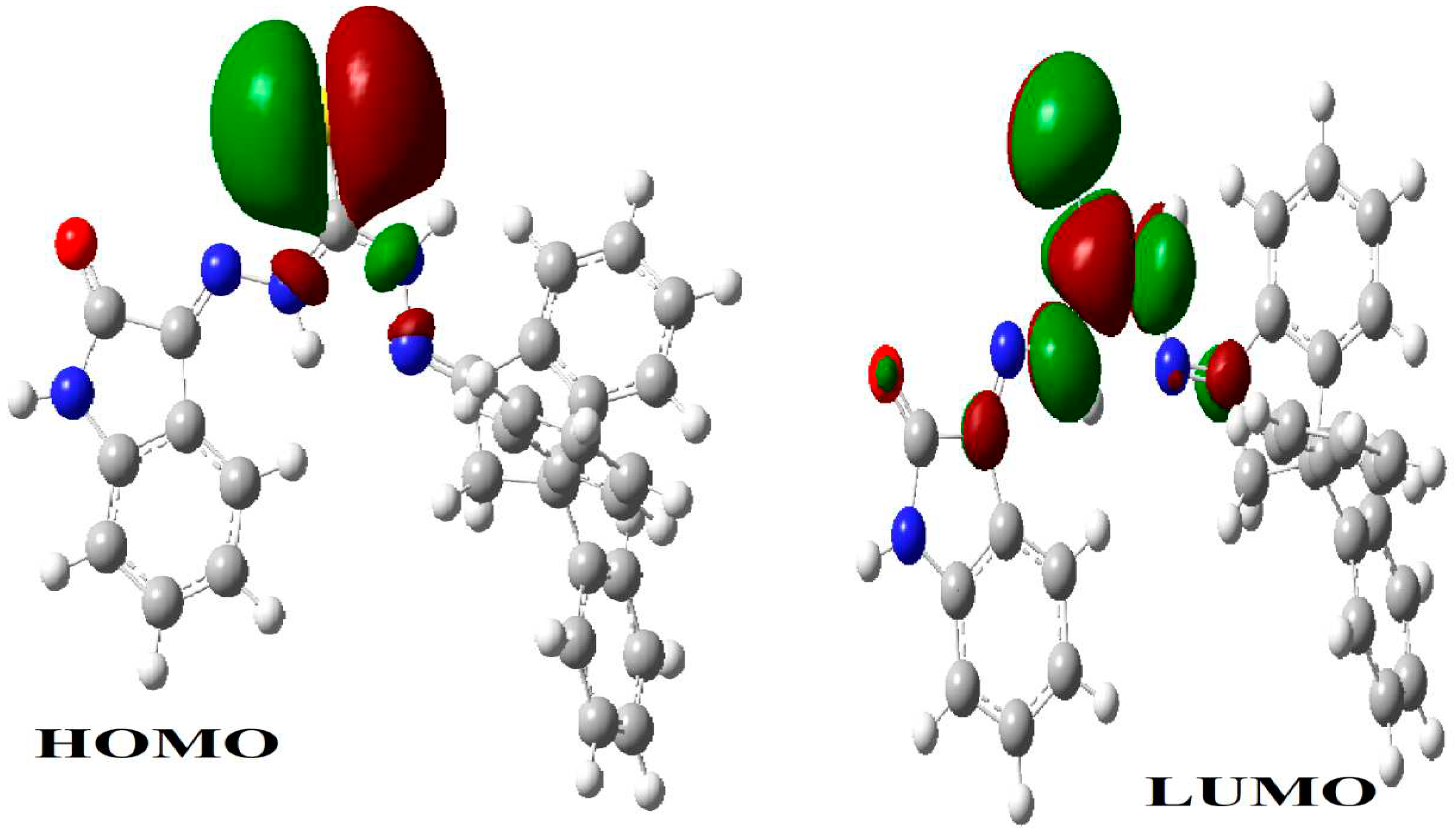
| Parameter | eV |
|---|---|
| EHOMO (eV) | -0.18396 |
| ELUMO (eV) | -0.08354 |
| µ(D) | -3.79687 |
| T.E (eV) | -1893.93 |
| ΔE (eV) | 0.10042 |
| χ (eV) | 0.13375 |
| η (eV) | 0.05021 |
| σ(eV) -1 | 19.91635 |
| Pi (eV) | -0.13375 |
| S (eV)-1 | 9.958176 |
| ω(eV) | 0.17814243 |
| ΔNmax | 2.66381199 |
| Compound | TG range朱(°C) | DTGmax朱(°C) | n* | Mass Loss Total mass Loss Calcd (Estim) % | Assignment | residue |
|---|---|---|---|---|---|---|
| H2L | 40–150朱150-250朱250-400 | 75朱200朱300 | 1朱1朱1 | 15.82 (15.56)朱34.51 (33.93) 99.19 (97.99)朱48.86 (35.61) | - Loss of C6H6朱- Loss of C9 H7N2S朱-Loss of C15H10N3O | -------- |
| [Co(H2L)2].Cl2 | 50-150朱150-250朱250-600朱600-1000 | 110朱200朱400朱800 | 1朱1朱1 | 7.16 (6.87)朱22.63 (22.31)93.37 ( 92.31)朱45.80 (44.98)朱17.78 (18.15) | - Loss of 2HCl, H2O and C2H2.朱- Loss ofC12H10N5S.朱- Loss of C33H20 N5S朱- Loss of C13H10 | CoO |
| [Ni(H2L)2].Cl2 | 40-160朱160-280朱280-650朱650-1000 | 110朱220朱480朱800 | 1朱1朱1朱1 | 11.76. (11.45)朱33.27 (32.76)朱26.90 (26.31)93.46 (91.39)朱21.53 (20.87) | - Loss of 2HCl, H2O and C3H5.朱- Loss of C21H17.N5S朱- Loss of C18H11N3S.朱- Loss of C18H9N2朱 | NiO |
| [Fe(H2L)2].Cl3 | 50-280朱280-570朱570-800朱800-1000 | 160朱320朱600朱850 | 1朱1朱1朱1 | 19.69 (20.32)朱28.80 (28.64)朱35.76 (35.72)94.22 (93.13)朱9.97 (8.45) | - Loss of 3HCl and C9H10.朱- Loss of C18H12N5 S.朱- Loss of C24H13N5S and 1/2 O.朱- Loss of C9H8. | ½Fe2O3 |
| [Cu(H2L)2].Cl2 | 40-250朱250-560朱560-750朱750-1000 | 170朱350朱660朱880 | 1朱1朱1朱1 | 13.72 (12.66)朱36.05 (35.72)朱33.42 (33.81) 93.91 (93.06)朱10.72 (10.84) | - Loss of H2O, 2HCl and C5H4.朱- Loss of C24H15N5S.朱- Loss of C23H12N4S.朱- Loss of C8H11N | CuO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





