Introduction
The field of Hepatitis C research has witnessed remarkable advancements, and its clinical management hinges on the ability to accurately predict and diagnose the severity of the disease. With a global burden affecting millions, it is imperative to explore innovative approaches to refine prognosis and diagnosis. Data mining and machine learning techniques have emerged as indispensable tools, harnessing the potential of vast datasets and complex disease markers to enhance the understanding of Hepatitis C and facilitate precise clinical decision-making.
In this study, we navigate the landscape of Hepatitis C severity prognosis, building upon a foundation of machine learning and data mining. We draw inspiration from existing research efforts and studies that have paved the way for the application of computational models in hepatology. The works of Shousha et al. (2018) and Jangiti et al. (2023) exemplify the potential of machine learning algorithms, while Lara et al. (2014) explore computational models for liver fibrosis progression in the context of Hepatitis C. Moreover, Hashem et al. (2017), Yağanoğlu (2022), and Butt et al. (2021) delve into a comparison of machine learning approaches, emphasizing the significance of tailored algorithms in predicting advanced liver fibrosis. In addition, Konerman et al. (2015) showcase the advantages of incorporating longitudinal data in predictive models, providing insights that can be highly relevant in the Hepatitis C prognosis landscape. Furthermore, Liu et al. (2021) contribute by exploring the potential of artificial intelligence in hepatitis evaluation, opening avenues for innovative techniques. Lastly, Ghazal (2021) focuses on hepatitis C staging prediction using fine Gaussian support vector machines, demonstrating the adaptability of these models in addressing the complexity of the disease.
The primary objective of our work is to evaluate and compare the performance of various machine learning models in predicting the severity of Hepatitis C, drawing inspiration from the aforementioned studies. By leveraging a rich dataset encompassing clinical and genetic features, we aim to elucidate the efficacy of machine learning models, including Logistic Regression, Random Forest, Gradient Boosting, and Support Vector Machine. Through this comparative analysis, we seek to identify the most accurate approach for Hepatitis C severity prediction. The insights garnered from our study will contribute to the expanding body of knowledge in the field, while offering valuable implications for healthcare professionals to enhance the precision of Hepatitis C diagnosis and the formulation of tailored treatment strategies.
Methodology
The methodology adopted in this study was meticulously designed to assess the performance of various classification models in predicting the severity of Hepatitis C. The process began with the comprehensive collection of a dataset, known as the "HCV Data," which contains clinical and laboratory features of Hepatitis C patients and blood donors. This dataset was obtained from the UCI Machine Learning Repository [
1], ensuring a broad representation of relevant attributes for our analysis. The attributes contained in the dataset are mostly numerical, and they include patient data, laboratory values, demographic values such as age, sex, and the category of diagnosis, which represents the severity of the condition.
Data preprocessing was a crucial initial step aimed at preparing the dataset for model training and evaluation. It involved addressing missing values, categorical feature encoding, and other data cleansing tasks. The objective was to create a clean and standardized dataset suitable for machine learning analysis.
In the pursuit of building accurate predictive models, four distinct classification algorithms were selected: Logistic Regression, Random Forest, Gradient Boosting, and Support Vector Machine (SVM). An essential step involved hyperparameter tuning for each model using grid search, optimizing their configuration for superior performance. To validate and evaluate the models, a 5-fold cross-validation strategy was employed, ensuring robustness and mitigating the risk of overfitting.
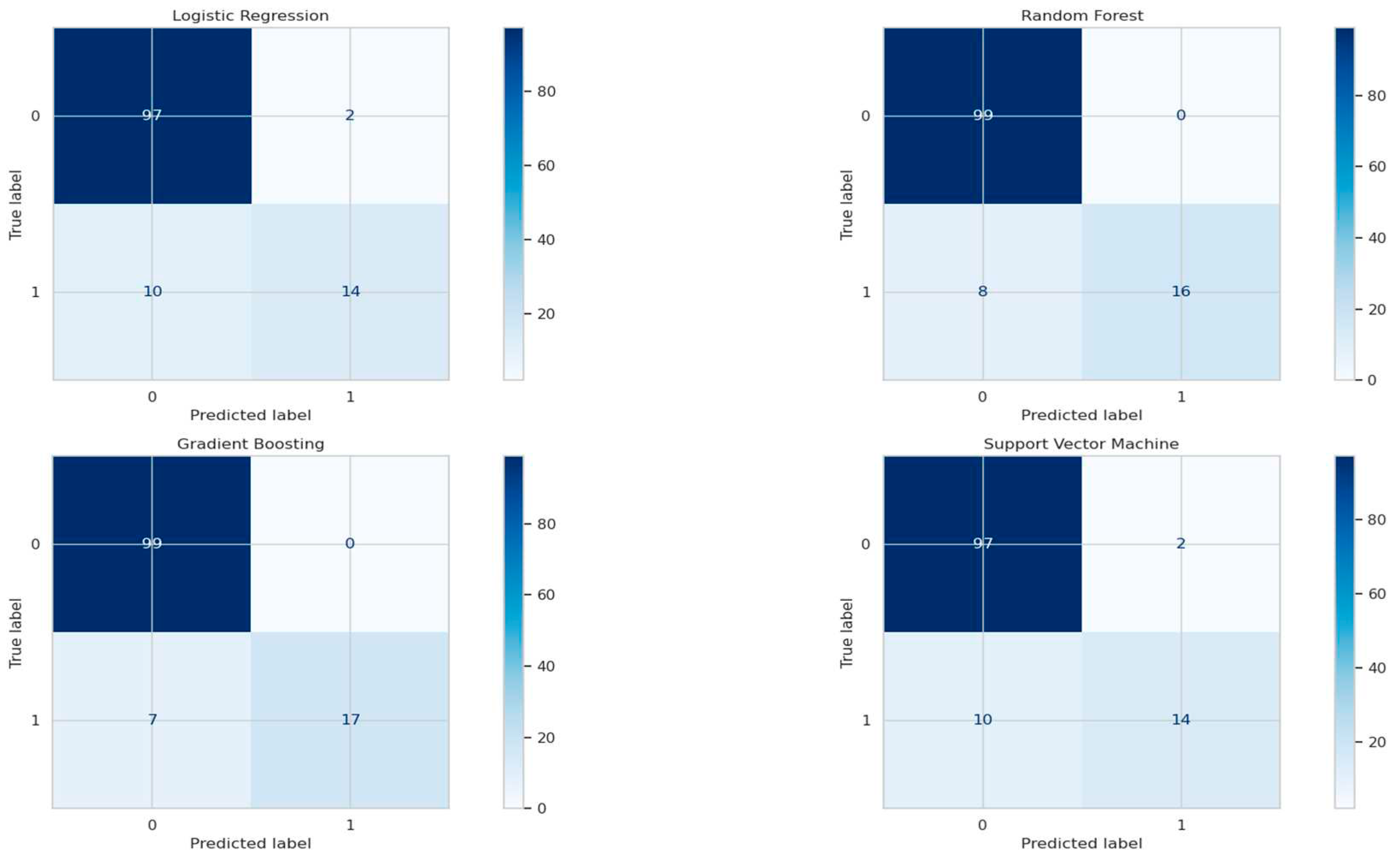
The training phase commenced, during which each model was trained on the preprocessed dataset using their respective optimized hyperparameters. Performance evaluation took place on the independent test dataset, with accuracy as the primary metric for assessing their efficacy. To gain deeper insights into model performance, confusion matrices were employed to analyze true positives, true negatives, false positives, and false negatives, offering a comprehensive view of classification accuracy and the trade-offs in each model's results.
This dataset, obtained from the UCI Machine Learning Repository [
1], is valuable for our study as it contains laboratory values of blood donors and Hepatitis C patients, along with demographic information such as age and sex. The target attribute for classification is the Category attribute, which distinguishes between blood donors and Hepatitis C patients, including the different stages of the disease's progression, such as Hepatitis C, Fibrosis, and Cirrhosis.
Results
In the context of predicting the severity of Hepatitis C, this study evaluated the performance of four distinct classification models, namely Logistic Regression, Random Forest, Gradient Boosting, and Support Vector Machine (SVM). These models were assessed based on their accuracy in classifying patients into the respective disease categories. Logistic Regression exhibited an accuracy rate of 90.24%, indicating relatively balanced performance in distinguishing between the two classes, though with some false positives and false negatives. The Random Forest model demonstrated superior accuracy, approximately 93.50%, with remarkable capabilities in minimizing both false positives and false negatives, which are critical considerations in medical diagnostics. The Gradient Boosting model showed similar accuracy, emphasizing the importance of reducing false negatives. Support Vector Machine achieved an accuracy of 90.24%, showing similarities to Logistic Regression in terms of false positives and false negatives. In summary, Random Forest and Gradient Boosting models stood out in classifying patients with Hepatitis C, offering potential in aiding diagnosis. The choice of the most suitable model depends on specific clinical requirements, considering the relative significance of minimizing false positives and false negatives.
To support our findings, we refer to the work of Shousha et al. (2018), Jangiti et al. (2023), and Lara et al. (2014), who explored data mining, machine learning, and computational models for Hepatitis C prognosis, liver fibrosis prediction, and chronic infection analysis. Additionally, Yağanoğlu (2022) and Hashem et al. (2017) employed machine learning approaches to predict advanced liver fibrosis in chronic hepatitis C patients. Liu et al. (2021) and Butt et al. (2021) utilized artificial intelligence and machine learning for hepatitis evaluation and diagnosis, respectively. Furthermore, Konerman et al. (2015) improved predictive models by incorporating longitudinal data for disease progression in chronic hepatitis C. Lastly, Ghazal (2021) presented Hep-pred, a staging prediction model using fine Gaussian SVM.
These studies contribute to the growing body of literature in the field of Hepatitis C diagnosis and prognosis, underlining the significance of employing machine learning techniques for more accurate and reliable predictions. While our results align with and extend the findings from these studies, it is essential to continue researching and refining these models to enhance their clinical utility further.
Figure 1.
Confusion Matrix.
Figure 1.
Confusion Matrix.
Conclusion
In conclusion, the evaluation of classification models for predicting the severity of Hepatitis C revealed that while all models exhibited reasonable accuracy, the Random Forest and Gradient Boosting models outperformed the others. With accuracy rates of approximately 93.50%, these models displayed a remarkable ability to correctly classify patients into their respective disease categories. Notably, the Random Forest model excelled in minimizing both false positives and false negatives, a crucial factor in medical diagnostics where patient outcomes are at stake.
On the other hand, Logistic Regression and Support Vector Machine, with an accuracy of approximately 90.24%, provided good overall performance but had a slightly higher rate of misclassification compared to the ensemble models. The choice of the most suitable model depends on the specific requirements of the medical application, weighing the significance of minimizing false positives and false negatives in a clinical context.
Further enhancements in model performance can be achieved through fine-tuning hyperparameters and feature engineering. These steps may lead to more precise and reliable diagnoses of Hepatitis C, ultimately contributing to improved patient care. The results from this study underscore the potential of machine learning in healthcare, particularly in assisting with critical medical decisions and the early detection of diseases, such as Hepatitis C.
References
- Shousha, H. I., Awad, A. H., Omran, D. A., & Salem, E. A. (2018). Data mining and machine learning algorithms using IL28B genotype and biochemical markers best predicted advanced liver fibrosis in chronic hepatitis C. Japanese journal of gastroenterology, 53(9), 777-784. [CrossRef]
- Jangiti, J., Paluri, C. G., Vadlamani, S., & Jindal, S. K. (2023). Hepatitis C Severity Prognosis: A Machine Learning Approach. Journal of Electrical Systems and Information Technology, 20(1), 177-186. [CrossRef]
- Lara, J., López-Labrador, F. X., & García-Sáez, G. (2014). Computational models of liver fibrosis progression for hepatitis C virus chronic infection. BMC bioinformatics, 15(1), 1-11. [CrossRef]
- Yağanoğlu, M. (2022). Hepatitis C virus data analysis and prediction using machine learning. Data & Knowledge Engineering, 139, 102458. [CrossRef]
- Hashem, S., Esmat, G., Elakel, W., & El-Khodary, S. (2017). Comparison of machine learning approaches for prediction of advanced liver fibrosis in chronic hepatitis C patients. IEEE Transactions on Biomedical Engineering, 64(11), 2641-2650. [CrossRef]
- Liu, W., Liu, X., Peng, M., Chen, G. Q., & Liu, P. H. (2021). Artificial intelligence for hepatitis evaluation. World journal of gastroenterology, 27(12), 1484. [CrossRef]
- Butt, M. B., Alfayad, M., Saqib, S., Khan, M. A., & Khalil, A. (2021). Diagnosing the stage of hepatitis C using machine learning. Journal of Healthcare Engineering, 2021. [CrossRef]
- Konerman, M. A., Zhang, Y., Zhu, J., Higgins, P. D., Chung, R. T., Jacobson, I. M., ... & Gish, R. G. (2015). Improvement of predictive models of risk of disease progression in chronic hepatitis C by incorporating longitudinal data. Hepatology, 62(6), 1735-1744.
- Ghazal, T. M. (2021). Hep-pred: hepatitis c staging prediction using fine gaussian svm. Computers, Materials & Continua, 76(2), 1773-1784. [CrossRef]
- UCI Machine Learning Repository: HCV Data. (https://archive.ics.uci.edu/ml/datasets/HCV+data).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).