1. Introduction
Due to recent advancements and availability of anti-retroviral therapy (ART), viral suppression has been achieved in a majority of HIV
+ patients in the United States [
1]. The use of ART to attain viral suppression has resulted in controlled viremia (mainly undetectable viral load), restored CD4 T cell counts, decreased risk of opportunistic infections and increased survival and life expectancy among HIV-1 infected patients [
2,
3]. Despite the success of ART in reducing viral loads to undetectable levels in HIV
+ patients, HIV-1 eradication has not been achieved and the viral genome persists in the reservoirs. Moreover, these HIV
+ patients exhibited a reduction in terminally differentiated T cells and HIV-specific CD8 T cell responses and improvement in CD4 T cell functions, indicating a decline of exhaustion/senescence of T cells due to viral suppression by long-term ART [
4,
5].
Because of a high rate of HIV-1 replication by the error-prone reverse transcriptase enzyme in HIV
+ patients with uncontrolled viremia, mutants rapidly accumulate over the course of infection leading to an increase in viral genetic diversity [
6]. While the immune system mounts a robust response, immunological-escape mutants arise during infection leading to persistence of HIV-1 infection reservoirs in patients [
7]. These HIV-1 reservoirs are established early after infection [
8] in quiescent cells such as resting memory CD4+ T cells, microglia cells, and monocytes/macrophages [
9]. These HIV-1 reservoirs persist for the lifetime [
10] of HIV
+ patients on ART but the mechanisms of residual HIV-1 maintenance are not known. The possibility exists that HIV-1 regulatory protein, Tat, which is essential for viral replication, may play a role in HIV-1 maintenance in HIV
+ older patients on ART that may be under differential selective pressure due to immunosenescence. Therefore, characterization of the molecular, biological, and immunological features of
tat gene from HIV
+ older patients with controlled viremia on long term ART may provide important information for the development of curative or therapeutic strategies.
The HIV-1 regulatory protein, Tat, is a multifunctional protein that facilitates HIV-1 infectivity through both the control of viral transcription and posttranscriptional regulation of viral and cellular gene expression [
11]. Tat is a nuclear protein that increases viral transcription through its interaction with the TAR element in the 5’ stem-loop RNA structure of HIV-1 LTR [
12]. Tat recruits P-TEFb complex to the nascent TAR RNA, activating RNA polymerase-II leading to significant increases in the transcriptional elongation of viral mRNA, facilitating HIV-1 replication[
13,
14]. Additionally, Tat acts as a posttranscriptional regulator of viral and cellular gene expression to create optimal conditions for HIV-1 replication[
11]. For example, Tat increases the transcription of chemokines [
15] and chemokine receptor CXCR4 [
16] promoting viral infectivity and disease progression. Moreover, Tat protein is secreted by infected cells, resulting in extracellular accumulation and internalization by neighboring cells leading to altered gene expression [
17]. Extracellular Tat has been found to contribute to neuropathogenesis in HIV-1-associated neurocognitive disorders[
18] and HIV-1-associated cardiovascular complications [
19]. More importantly, even with successful viral suppression due to ART, HIV
+ patients’ cells still produce and secrete Tat protein [
20], suggesting the vital role of Tat in regulation of viral replication and cellular gene expression. Therefore, we sought to characterize HIV-1
tat sequences from HIV
+ older patients who have achieved controlled viremia (undetectable viral load) and improved CD4 T cell counts due to long-term ART to understand Tat’s role in viral persistence and its interactions with the aging immune systems.
We show that HIV-1 tat sequences from 20 HIV+ older patients on long-term ART, who have achieved undetectable viral load and restored CD4 T cell counts, exhibited a low degree of viral heterogeneity and lower estimates of viral diversity. The majority of patients’ sequences had dN/dS of <1, suggesting weak directional selective pressure probably due to immune aging of the older patients. More importantly, the functional domains required for Tat activity, including transactivation, TAR binding, and nuclear localization were conserved, while previously identified CTL epitopes were variable, suggesting immune escape mutants.
2. Materials and Method
2.1. Study Design, Human Subjects, and PBMC Preparation
This was a cross-sectional analysis using PBMC and plasma samples obtained from 20 HIV
+ patients in 2017 in Tucson, Arizona. This study was approved by the Institutional Review Board of the University of Arizona, Tucson. Informed and signed consent forms were obtained from each patient and data was processed using unique identifiers to ensure confidentiality. This study included 20 HIV
+ older patients with ages ranging from 39-81 years (average age: 56 years) with undetectable viral load ranging from <20 to 105 (mostly <20) HIV RNA copies/mL and CD4 T cell counts ranging from 248 to 1469 (average: 626 cells/mL). These patients received health care and long-term ART at the Petersen HIV-1 Clinic, Infectious Disease Division, Banner University of Arizona Medical Center.
Table 1 contains the demographic, clinical, and laboratory data from this cohort of HIV-1-infected patients. Blood was collected in 10mL sodium heparin Vacutainer tubes (BD, Sunnyvale, CA) for each patient and was processed to isolate peripheral blood mononuclear cells (PBMCs) and plasma. Samples were cryopreserved by University of Arizona Biorepository Laboratory for future analysis.
Table 1.
Patient Demographic, Clinical, and Laboratory Parameters of HIV-1 infected older patients on long term ART.
Table 1.
Patient Demographic, Clinical, and Laboratory Parameters of HIV-1 infected older patients on long term ART.
| |
Patient |
Age (Yrs) |
Sex |
Race |
Ethnicity |
Years since Dx |
Dx Aids |
Viral Load (copies/mL) |
CD4+ (cells/mL) |
| 1 |
AK 8 |
47 |
F |
B |
NH |
10 |
Yes |
<20 |
1088 |
| 2 |
AK 16 |
52 |
M |
O |
NH |
25 |
Yes |
<20 |
770 |
| 3 |
AK 19 |
81 |
M |
W |
NH |
8 |
No |
N/A |
778 |
| 4 |
AK 24 |
44 |
M |
W |
NH |
13 |
No |
<20 |
653 |
| 5 |
AK 36 |
53 |
M |
W |
H |
26 |
No |
<20 |
583 |
| 6 |
AK 55 |
49 |
M |
W |
NH |
4 |
Yes |
105 |
444 |
| 7 |
AK 57 |
39 |
F |
B |
NH |
1 |
No |
<20 |
1029 |
| 8 |
AK 58 |
63 |
M |
W |
NH |
29 |
Yes |
<20 |
1469 |
| 9 |
AK 68 |
53 |
F |
W |
NH |
25 |
Yes |
<20 |
374 |
| 10 |
AK 71 |
56 |
M |
W |
NH |
14 |
Yes |
<20 |
452 |
| 11 |
AK 78 |
49 |
M |
W |
NH |
10 |
No |
<20 |
696 |
| 12 |
AK 83 |
53 |
M |
W |
NH |
13 |
No |
29 |
240 |
| 13 |
AK 92 |
72 |
M |
W |
H |
9 |
Yes |
58 |
638 |
| 14 |
AK 96 |
75 |
M |
W |
NH |
34 |
No |
N/A |
N/A |
| 15 |
AK 97 |
67 |
M |
W |
NH |
16 |
Yes |
<20 |
561 |
| 16 |
AK 99 |
50 |
M |
A |
H |
9 |
Yes |
<20 |
248 |
| 17 |
AK 100 |
54 |
M |
W |
H |
17 |
Yes |
<20 |
342 |
| 18 |
AK 110 |
57 |
M |
W |
NH |
5 |
Yes |
<20 |
470 |
| 19 |
AK 125 |
61 |
M |
W |
NH |
32 |
Yes |
76 |
431 |
| 20 |
AK 187 |
46 |
M |
W |
NH |
11 |
No |
<20 |
985 |
| |
Average |
56 |
|
|
|
15.55 |
|
|
|
|
Abbreviations: M: Male, F: Female, W: White, MA: Mexican American, B: Black, O: Other, H: Hispanic, NH: Non-Hispanic, N/A: Not available |
2.2. PBMC DNA Isolation, PCR Amplification, Cloning and Sequencing
The Purelink Genomic DNA Kit (Invitrogen Inc.) was used to extract the DNA from cryopreserved peripheral blood mononuclear cell (PBMC) samples (10x106) according to the manufacturer’s protocol. PBMC samples were resuspended in a phosphate buffered saline (PBS) and incubated with equal volumes of Proteinase K and RNase A and incubated at room temperature for 2 minutes. Two hundred μL of the PureLink Genomic Lysis/Binding buffer was added to each sample and incubated at 55°C for 10 minutes followed by addition of 200μl of 100% ethanol and the sample was transferred to a PureLink Spin Column and centrifuged. The eluate was transferred back into the column to maximize DNA capture. Wash Buffer 1 and Wash Buffer 2 (500μL) were added to the column and centrifuged at 10,000xg. 50uL of the PureLink Genomic Elution Buffer were added to the column, incubated for 1 minute and centrifuged at maximum speed for 1.5 minutes followed by a second elution with 50μL. The DNA concentration was determined using a Thermo Scientific Spectrophotometer NanoDrop and stored at -20°C for PCR amplification use.
A 559-bp fragment encompassing both HIV-1 tat and vpu genes were amplified from 20 HIV+ patients’ PBMC DNA using a nested two-step polymerase chain reaction (PCR) . Primers were designed using the HIV-1 NL4-3 sequence which belongs to subtype B of HIV-1. The two outer primers for the first PCR were TAT-1 (5’CAACTGCTGTTTATCCATTTCAGAA3’) and VPU-2 (5’ATATGCTTTAGCATCTGATGCACAA3’). Each outer PCR was performed in 25µl reactions containing 250ng PBMC DNA, 2.5µl 10xTaq Buffer, 2.5mM dNTPs, 2mM MgCl2, .5µM of each primer and 1.5 units of Takara Ex Taq DNA polymerase. The PCR was carried for 40 cycles with an initial denaturation at 95°C for 2 min followed by 98°C for 10s, 52°C for 45s, 72°C for 2 min, and a final extension phase at 72°C for 8 min. The outer PCR products were then purified using the Invitrogen Purelink PCR purification kit according to the manufacturer’s protocol. Concentrations of the purified PCR product were determined using a Thermo Scientific Spectrophotometer Nanodrop. Next, another PCR reaction was completed using two inner primers, TAT-3 (5’GTGTCGACATAGCAGAATAGGC3’) and VPU-4 (5’CCATAATAGACTTGTGACCCACA3’). The inner PCR conditions were carried for 35 cycles with an initial denaturation at 95°C for 2 min, followed by 98°C for 30s, 58°C for 45s, 72°C for 2 min, and an extension period at 72°C for 8 min. For both PCR reactions, sterile water was used as a negative control and HIV-1 NL4-3 was used as a positive control. PCR products were electrophoresed on 1.4% agarose gels and visualized using ethidium bromide staining under UV light to confirm the correct size band of ~559 base pairs and confirm the negative control was free from contamination. All samples, reagents and PCR products were stored separately to prevent PCR contamination.
Inner PCR products with the correct sized DNA fragments (~559 base pairs) were cloned into the pCR 4-TOPO TA Vector, ligated, and transformed into chemically competent Escherichia coli cells according to the manufacturer’s protocol (Invitrogen, Inc.). Bacterial colonies were grown overnight, and plasmid DNA was extracted using Miniprep procedure. Plasmid DNAs were screened for the presence of recombinants by restriction enzyme digestion (EcoR1). To confirm the presence of correct sized recombinants, gel electrophoresis was conducted on a 1.4% agarose gel and clones with the correct sized recombinants were selected to be sequenced. An average of 15 clones per patient, ranging between 6 and 41 clones, including the positive control HIV-1 NL4-3 obtained from multiple independent PCRs were sequenced by Sanger DNA Sequencing method at the University of Arizona Genetics Core (Applied Biosystems 3730 DNA Analyzers)
2.3. Sequence Analysis
Nucleotide sequences of the HIV-1
tat gene (216 bases) encompassing Exon 1 (coding region) from 20 HIV
+ patients were analyzed using the CLC Main work Bench 7 (Version 7.7.3 by Qiagen). Nucleotide sequences were assembled, analyzed, and translated into their deduced amino acid sequences and aligned using HIV-1 NL4-3 as the references sequence. A phylogenetic tree was created using the neighbor-joining method and the nucleotide distance was measured using Jukes-Cantor. The neighbor-joining tree was bootstrapped 1,000 times to ensure the validity of the phylogenetic tree. The minimum, median and maximum for both nucleotides and amino acids distances were calculated for each patients using the Jukes-Cantor model [
21]. The Jukes-Cantor model assumes equal base and amino acid frequencies and equal substitution rates. Population genetics was then used to assess the variation of the HIV-1
tat gene evolution within 20 HIV+ patients. Genetic diversity is defined as y = 2N
eμ, where N
e is the effective population size and μ is the per nucleotide mutation rate per generation. Genetic diversity was calculated using the Watterson 1975 equation 1.4a estimate based on segregating sites and the Nei 1987 equation 10.3 estimate assuming variable population size using the DnaSP DNA sequence Polymorphism 5.10.01 software[
22,
23]. The ratio of nonsynonymous (dN) to synonymous (dS) substitutions was estimated to determine the evolutionary processes acting on the HIV-1
tat gene in HIV-1-infected older patients using the Nei and Gojobori 1986 model[
24]. This model considers the codon instead of the nucleotide as the unit of evolution and provides two separate estimates, one for diversity and one for divergence.
Sequence Accession number: The sequences were submitted to GenBank and accession numbers provided are OR265643 - OR265939.
3. Results
3.1. Phylogenetic Analysis of HIV-1 tat Sequences from HIV+ Older Patients on Long- Term ART
The
tat gene was amplified using multiple independent PCR amplifications from the PBMC DNA of 20 HIV
+ patients with controlled viremia on long term ART yielding a total of 297 sequences. Between 6-41 sequences per patient were included in the phylogenetic analysis with an average of 15 sequences per patient. The neighbor-joining tree method was used to create a phylogenetic tree using 297
tat gene nucleotide sequences from 20 HIV
+ patients’ sequences. The HIV-1 NL4-3 sequence was used as the reference sequence to provide a root base of the phylogenetic tree.
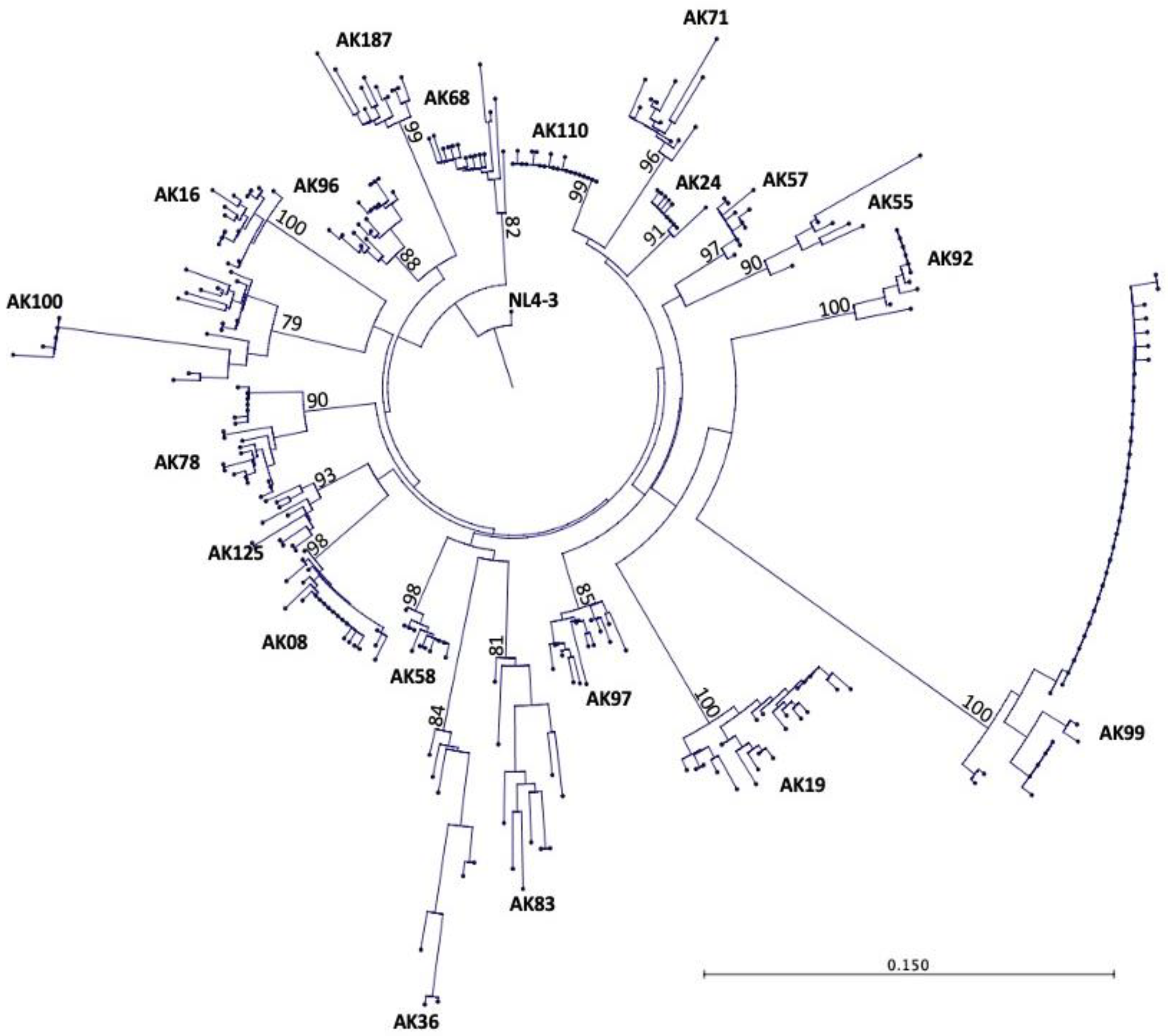
Figure 1 shoes the phylogenetic tree with each terminal node representing one
tat sequence. The phylogenetic tree showed that the
tat gene in each HIV
+ patient has evolved separately and distinctly from other HIV
+ patients and the reference sequence, HIV-1 NL4-3. All HIV
+ patients’ tat sequences belonged to HIV-1 subtype/clade B sequences. These findings were validated by bootstrapping the data set 1,000 times, which showed high bootstrap values between 79-100% for the 20 patients’ sequences.
Figure 1.
Phylogenetic tree of 297 tat gene sequences from 20 HIV+ older patients on long term ART. Each terminal node on the phylogenetic tree represents one tat sequence. The numbers on the branches of the phylogenetic tree represent the percent occurrences of the branches over 1,000 bootstrap resampling’s of the data set. The high bootstrap values indicate that the data is strongly supported in the phylogenetic tree, which reveals that all 20 HIV+ patients were well discriminated and confined to their own subtrees, showing no PCR cross contamination.
Figure 1.
Phylogenetic tree of 297 tat gene sequences from 20 HIV+ older patients on long term ART. Each terminal node on the phylogenetic tree represents one tat sequence. The numbers on the branches of the phylogenetic tree represent the percent occurrences of the branches over 1,000 bootstrap resampling’s of the data set. The high bootstrap values indicate that the data is strongly supported in the phylogenetic tree, which reveals that all 20 HIV+ patients were well discriminated and confined to their own subtrees, showing no PCR cross contamination.
3.2. Coding Potential of tat Deduced Amino Acid Sequences from HIV+ Older Patients on Long-Term ART
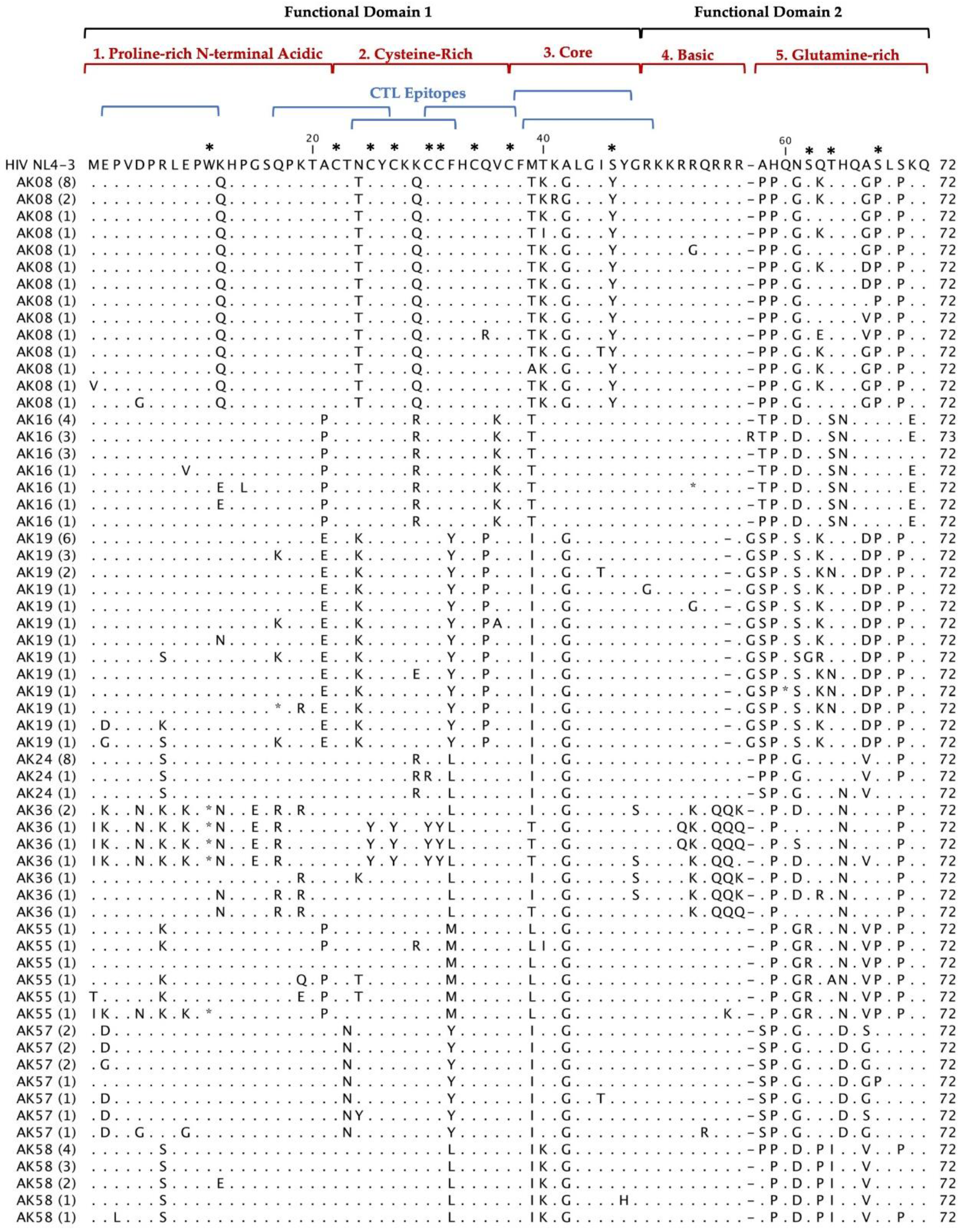
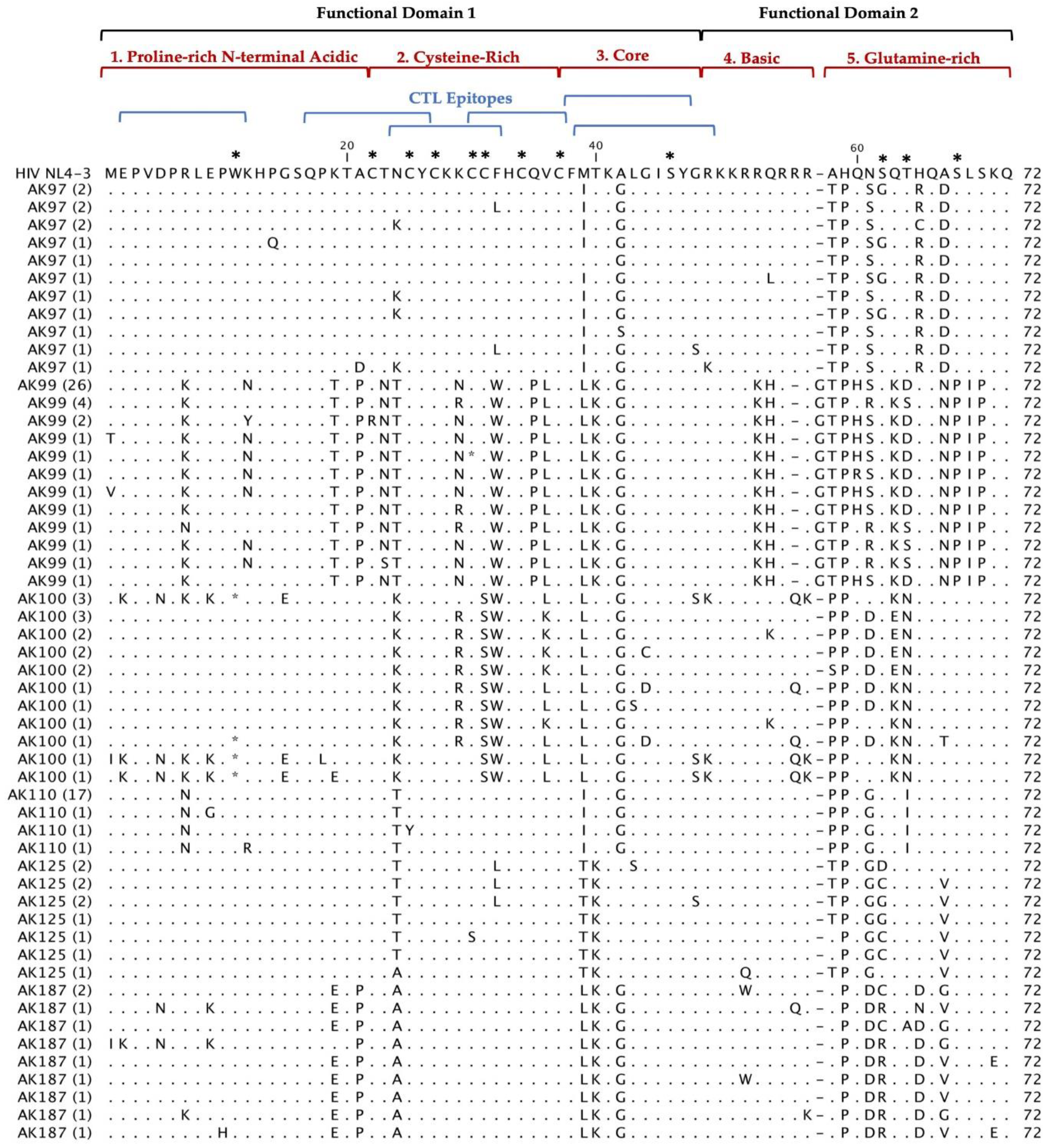
Multiple alignments of 297 Tat deduced amino acid sequences from 20 HIV
+ patients aligned with the reference HIV-1 NL4-3 sequences are shown in
Figure 2. The coding potential of the
tat gene was maintained in 269 (90.6%) sequences, with only 28 sequences carrying inactivating mutants. Most of the sequences that did not have an intact open reading frame came from three patients: AK36, AK83 and AK100. Most of the inactivating mutants (stop codons) were located in the first functional domain of Tat at position W11(tryptophan), an amino acid necessary for secretion of Tat which is typically conserved among in HIV
+ patients’ sequences [
25]. There was one insertion before position 58 found in our patients Tat sequences compared with HIV-1 NL4-3. In AK19, AK92, and AK99 contained an insertion of G (glycine) and AK16 contained an insertion of R (arginine). Furthermore, the deduced amino acid sequences exhibited patient-specific amino acid motifs exclusive to their respective patients, while other amino acid motifs were shared among sequences from multiple patients.
Figure 2.
Deduced Amino Acid Alignment of 297 tat gene of 72 amino acids from 20 HIV+ older patients with controlled viremia and improved CD4 T cell counts. Deduced amino acid sequences were aligned using HIV-1- NL4-3 strain as the reference sequence shown at the top of the alignment. Dots represent amino acid agreement with the reference sequence and substitutions are represented by their corresponding amino acid code. Asterisks represent stop codons. Dashes represent gaps in the amino acid sequence due to either addition or deletion of an amino acid. Above the alignment are lines representing the two functional domains of Tat exon I comprising of specific regions for Tat functions. In addition, the six blue lines represent CTL epitopes analyzed in the Tat sequence. Amino acids that are important for Tat function have stars above them at the top of the alignment.
Figure 2.
Deduced Amino Acid Alignment of 297 tat gene of 72 amino acids from 20 HIV+ older patients with controlled viremia and improved CD4 T cell counts. Deduced amino acid sequences were aligned using HIV-1- NL4-3 strain as the reference sequence shown at the top of the alignment. Dots represent amino acid agreement with the reference sequence and substitutions are represented by their corresponding amino acid code. Asterisks represent stop codons. Dashes represent gaps in the amino acid sequence due to either addition or deletion of an amino acid. Above the alignment are lines representing the two functional domains of Tat exon I comprising of specific regions for Tat functions. In addition, the six blue lines represent CTL epitopes analyzed in the Tat sequence. Amino acids that are important for Tat function have stars above them at the top of the alignment.
3.3. Variability of tat Sequences from HIV+ Older Patients on Long Term ART
To evaluate the genetic variability of
tat gene, both the nucleotide and deduced amino acid distances from 20 HIV
+ patients’ sequences were estimated. The minimum, median, and maximum nucleotide distance and deduced amino acid distance distances are shown in
Table 2. The average nucleotide sequence distance for the
tat gene ranged from 0.00% to 0.12% with a median of 0.03% among 20 HIV
+ patients’ sequences. AK99 had the largest nucleotide distance observed, 0.08% (median) and 0.24% (maximum). The average deduced amino acid sequence distance for the
tat gene ranged from 0.00% to 0.48%. The largest distance in deduced amino acid sequences was 0.48% observed in the same patient, AK99. Nucleotide and amino acid sequence distances were correlated with the CD4 T cell counts of the HIV
+ patients. We found a weak negative correlation between the nucleotide and amino acid distance and the CD4 T cell count. HIV-1-infected patients with CD4 T cell counts <500cells/mL tended to have greater nucleotide and amino acid distances compared to patients with CD4 T cell counts > 500cells/mL. There was no correlation found between the patients’ length of infection and distance of
tat sequences, including increasing age, likely because of controlled viremia due to ART.
Table 2.
Nucleotide and amino acid distances in tat sequences from HIV+ older patients on long term ART.
Table 2.
Nucleotide and amino acid distances in tat sequences from HIV+ older patients on long term ART.
| |
|
|
Nucleotides % |
Amino Acids % |
| |
Patient |
N |
Min |
Median |
Max |
Min |
Median |
Max |
| 1 |
AK 8 |
22 |
0 |
0.01 |
0.1 |
0 |
0.03 |
0.24 |
| 2 |
AK 16 |
14 |
0 |
0.01 |
0.1 |
0 |
0.01 |
0.18 |
| 3 |
AK 19 |
21 |
0 |
0.02 |
0.13 |
0 |
0.03 |
0.27 |
| 4 |
AK 24 |
10 |
0 |
0.01 |
0.08 |
0 |
0 |
0.17 |
| 5 |
AK 36 |
8 |
0 |
0.08 |
0.18 |
0 |
0.17 |
0.41 |
| 6 |
AK 55 |
6 |
0.01 |
0.05 |
0.17 |
0.03 |
0.07 |
0.27 |
| 7 |
AK 57 |
10 |
0 |
0.01 |
0.09 |
0 |
0.03 |
0.2 |
| 8 |
AK 58 |
11 |
0 |
0.01 |
0.08 |
0 |
0.03 |
0.18 |
| 9 |
AK 68 |
15 |
0 |
0.02 |
0.1 |
0 |
0.06 |
0.2 |
| 10 |
AK 71 |
11 |
0 |
0.03 |
0.1 |
0 |
0.04 |
0.17 |
| 11 |
AK 78 |
20 |
0 |
0.04 |
0.09 |
0 |
0.06 |
0.18 |
| 12 |
AK 83 |
10 |
0 |
0.07 |
0.17 |
0 |
0.13 |
0.37 |
| 13 |
AK 92 |
10 |
0 |
0.01 |
0.11 |
0 |
0 |
0.24 |
| 14 |
AK 96 |
16 |
0 |
0.02 |
0.08 |
0 |
0.04 |
0.18 |
| 15 |
AK 97 |
14 |
0 |
0.03 |
0.09 |
0 |
0.04 |
0.15 |
| 16 |
AK 99 |
41 |
0 |
0 |
0.24 |
0 |
0.01 |
0.48 |
| 17 |
AK 100 |
18 |
0 |
0.06 |
0.16 |
0 |
0.09 |
0.35 |
| 18 |
AK 110 |
20 |
0 |
0 |
0.07 |
0 |
0 |
0.13 |
| 19 |
AK 125 |
10 |
0 |
0.03 |
0.09 |
0 |
0.06 |
0.15 |
| 20 |
AK 187 |
10 |
0 |
0.03 |
0.11 |
0 |
0.07 |
0.22 |
| |
Average |
|
0 |
0.03 |
0.12 |
0 |
0.05 |
0.24 |
| N = Number of HIV-1 tat clones sequenced |
3.4. Dynamics of HIV-1 tat Gene Sequence Evolution in HIV-1-Infected Older Patients on Long Term ART
To determine the sequence evolution of the
tat gene in our patients, we used two phylogenetic estimators of genetic diversity: the Watterson model and the Nei Model. The Watterson model estimates genetic diversity based on segregating sites [
22] while the Nei model estimates genetic diversity assuming a variable population size. The genetic diversity parameter, θ, estimated as nucleotide substitutions per site per generation was estimated for each patient and shown in
Table 3. The level of genetic diversity estimated by the Watterson model from 20 HIV
+ patients
tat sequences ranged from 0.022 to 0.076 (average: 0.047) and by the Nei model ranged from 0.008 to 0.083 (average: 0.037). The estimates of genetic diversity by both the Watterson and Nei models indicate that the
tat gene sequences from our 20 HIV
+ patients had low estimates of genetic diversity. Estimates of genetic diversity for the
tat gene were correlated with the patient’s CD4 T cell counts. HIV
+ patients with CD4 T cell counts <500 cells/mL showed higher levels of genetic diversity (average: 0.054) when compared with patients with >500 CD4 T cell counts (average: 0.044). The data suggests that there were higher estimates of genetic diversity in the
tat sequences of HIV
+ patients who had less recovered CD4 T cell counts.
Table 3.
Estimates of genetic diversity of the tat gene sequences from HIV-1-Infected older patients on long term ART.
Table 3.
Estimates of genetic diversity of the tat gene sequences from HIV-1-Infected older patients on long term ART.
| |
Patient |
N |
C |
θW
|
θN
|
| 1 |
AK8 |
22 |
72 |
0.036 |
0.017 |
| 2 |
AK16 |
14 |
73 |
0.034 |
0.022 |
| 3 |
AK19 |
21 |
71 |
0.053 |
0.032 |
| 4 |
AK24 |
10 |
72 |
0.035 |
0.021 |
| 5 |
AK36 |
8 |
71 |
0.075 |
0.083 |
| 6 |
AK55 |
6 |
71 |
0.076 |
0.066 |
| 7 |
AK 57 |
10 |
72 |
0.036 |
0.023 |
| 8 |
AK58 |
11 |
71 |
0.032 |
0.021 |
| 9 |
AK68 |
15 |
71 |
0.049 |
0.031 |
| 10 |
AK 71 |
11 |
71 |
0.054 |
0.035 |
| 11 |
AK78 |
20 |
71 |
0.041 |
0.035 |
| 12 |
AK83 |
10 |
72 |
0.073 |
0.078 |
| 13 |
AK92 |
10 |
71 |
0.043 |
0.030 |
| 14 |
AK96 |
16 |
71 |
0.032 |
0.026 |
| 15 |
AK97 |
14 |
71 |
0.048 |
0.032 |
| 16 |
AK99 |
41 |
71 |
0.056 |
0.024 |
| 17 |
AK100 |
18 |
71 |
0.065 |
0.067 |
| 18 |
AK 110 |
20 |
71 |
0.022 |
0.008 |
| 19 |
AK125 |
10 |
71 |
0.041 |
0.039 |
| 20 |
AK187 |
10 |
71 |
0.049 |
0.041 |
| |
Average |
|
|
0.047 |
0.037 |
| N = Number of HIV-1 tat clones sequenced, C = Number of codons analyzed per patient, θW = Genetic Diversity estimated by the Watterson, θN = Genetic Diversity estimated by the Nei |
3.5. Selective Pressure on tat Sequences from HIV+ Patients on Long Term ART
The accumulation of non-synonymous and synonymous substitutions was estimated to provide insight into the selective pressure and drift on the HIV-1
tat gene in our HIV
+ patients. Nucleotide substitutions that result in amino acid substitutions are called nonsynonymous substitutions(dN), whereas nucleotide substitutions that do not result in a change in amino acid sequence are synonymous substitutions (dS). A dN/dS ratio>1, in which dN will be expected to be faster than dS, indicates the influence of positive selection where non-synonymous substitutions produces beneficial adaptive changes in the protein sequence [
26]. The Nei and Gojobori maximum likelihood model was used to determine the evolutionary process on the
tat gene in HIV-1-infected older patients [
24]. This model considers the codon as the unit of evolution and provides estimates for both diversity and for divergence shown in
Table 4. The ratios of dN/dS calculations show that 13 out of 20 patients’
tat sequences had ratios of <1 indicating a weak directional selection toward negative pressure, which is consistent with the findings of another study of Tat [
27].
Table 4.
dN/dS ratios in the tat sequences from HIV+ older patients on long term ART.
Table 4.
dN/dS ratios in the tat sequences from HIV+ older patients on long term ART.
| |
|
|
|
Nucleotide Diversity |
Nucleotide Divergence |
| |
Patient |
N |
C |
dN |
dS |
dN/dS |
dN |
dS |
dN/dS |
| 1 |
AK8 |
22 |
72 |
0.017 |
0.012 |
1.508 |
0.017 |
0.011 |
1.530 |
| 2 |
AK16 |
14 |
73 |
0.016 |
0.042 |
0.385 |
0.015 |
0.039 |
0.386 |
| 3 |
AK19 |
21 |
71 |
0.030 |
0.033 |
0.895 |
0.030 |
0.033 |
0.897 |
| 4 |
AK24 |
10 |
72 |
0.016 |
0.036 |
0.438 |
0.015 |
0.033 |
0.443 |
| 5 |
AK36 |
8 |
71 |
0.086 |
0.052 |
1.645 |
0.086 |
0.052 |
1.643 |
| 6 |
AK55 |
6 |
71 |
0.056 |
0.090 |
0.621 |
0.056 |
0.089 |
0.624 |
| 7 |
AK 57 |
10 |
72 |
0.024 |
0.016 |
1.509 |
0.022 |
0.014 |
1.540 |
| 8 |
AK58 |
11 |
71 |
0.017 |
0.035 |
0.497 |
0.017 |
0.035 |
0.496 |
| 9 |
AK68 |
15 |
71 |
0.027 |
0.044 |
0.613 |
0.027 |
0.044 |
0.614 |
| 10 |
AK 71 |
11 |
71 |
0.036 |
0.028 |
1.275 |
0.036 |
0.028 |
1.281 |
| 11 |
AK78 |
20 |
71 |
0.031 |
0.045 |
0.694 |
0.031 |
0.045 |
0.695 |
| 12 |
AK83 |
10 |
72 |
0.068 |
0.090 |
0.762 |
0.062 |
0.082 |
0.760 |
| 13 |
AK92 |
10 |
71 |
0.020 |
0.060 |
0.328 |
0.020 |
0.060 |
0.331 |
| 14 |
AK96 |
16 |
71 |
0.020 |
0.046 |
0.443 |
0.020 |
0.046 |
0.442 |
| 15 |
AK97 |
14 |
71 |
0.029 |
0.037 |
0.784 |
0.029 |
0.037 |
0.785 |
| 16 |
AK99 |
41 |
71 |
0.023 |
0.021 |
1.084 |
0.023 |
0.021 |
1.109 |
| 17 |
AK100 |
18 |
71 |
0.065 |
0.061 |
1.057 |
0.065 |
0.061 |
1.067 |
| 18 |
AK 110 |
20 |
71 |
0.009 |
0.004 |
2.246 |
0.009 |
0.004 |
2.260 |
| 19 |
AK125 |
10 |
71 |
0.037 |
0.043 |
0.851 |
0.037 |
0.043 |
0.855 |
| 20 |
AK187 |
10 |
71 |
0.028 |
0.083 |
0.337 |
0.028 |
0.083 |
0.338 |
| |
Average |
|
|
0.033 |
0.044 |
0.899 |
0.032 |
0.043 |
0.905 |
N = Number of HIV-1 tat clones sequenced, C = Number of codons analyzed per patient
dN = Number of nonsynonymous substitutions per nonsynonymous site
dS = Number of synonymous substitutions per synonymous site |
3.6. Functional Domains Analysis in Tat Deduced Amino Acid Sequences from HIV+ Older Patients on Long Term ART
We analyzed the functional domains of Tat in a total of 297 deduced amino sequences from 20 HIV
+ older patients on long term ART. The first coding exon of Tat consists of 71 amino acids that form five regions that are responsible for Tat functioning. These regions are 1) the proline-rich N-terminal acidic domain (amino acids 1-21), 2) a cysteine-rich region (amino acids 22-37), 3) a core region (amino acids 38-47) containing hydrophobic amino acids, 4) a basic RNA binding region (amino acids 48-57) containing six arginine and two lysine (features of RNA-binding proteins) and 5) a glutamine rich region at the C-terminus (amino acids 58-71) [
28]. The first functional domain is important for transactivation of TAR and consists of the amino-terminal, cysteine-rich and core regions of amino acids 1-47 [
29]. The second functional domain consist of the basic and glutamine rich regions that are required for RNA binding, nuclear localization, and stabilization of the Tat protein [
30]. Our data show that each patient had unique patterns of amino acid sequences while preserving the important amino acids in each of the functional domains (
Figure 2).
3.6.1. Proline-Rich N-Terminal Acidic Domain:
The proline-rich N-terminal acidic domain includes amino acids 1-21 of Tat and is responsible in mediating LTR transactivation[
31]. The proline (P) at positions 3, 6, 10, 14, and 18 were highly conserved (98.3%) in our Tat sequences from 20 HIV
+ older patients, except 5 sequences that had substitutions P10H (patient AK187), P10L (AK16), P14Q (AK97), P18Q (AK97), P18G (AK71). The proline-rich N-terminal acidic domain works in conjunction with the cysteine-rich and core domains to interact via CYCT1 to activate LTR transactivation [
32]. A well-conserved amino acid is a tryptophan residue at position 11 that is necessary for efficient secretion of Tat [
25]. This is consistent with findings from this study, which had W11 conserved in 93% of patients’ Tat sequences. There were 20 sequences with premature stop codons at W11, mostly in AK36, AK83 and AK100. Additionally, amino acids R7, K19, and A21 displayed greater variability and contained substitutions in multiple patients’ sequences. Substitutions identified included R7S, R7K, R7E and R7N in several sequences from 11 patients. In addition, K19R, K19Q, K19E, K19T substitutions were found in 10 patients. Finally, substitutions identified in 8 patients including A21P, A21E, A21N, and A21D. Most of these substitutions were not in the important motifs responsible for Tat function, although some of the substitutions were with conservative amino acids.
3.6.2. Cysteine Rich Region
The cysteine-rich region (aa 22-37) of the Tat protein contains a highly conserved series of cysteine (C) residues that form intra-molecular disulfide bonds. Oxidation of sulfhydryl groups result in changes in the structure and biological activity of Tat [
33]. Studies indicate that mutations in these cysteine residues abolish the activity of the Tat protein [
28,
29]. Comparison of our patients’ 297 Tat sequences with HIV-1 NL 4-3 sequence shows that the cysteine-rich region at positions 22, 25, 27, 30, 34, and 37 were highly conserved (98% to 100%) in all 20 HIV
+ older patients’ (Fig. 2). At positions 34 and 37, there was 100% conservation of cysteine residues. Positions 25, 27, and 30, had 98% conservation of the cysteine residues. At C22, there was 99% conservation of cysteine, with AK99 having a substitution of C22R. Patient AK100 had a substitution C31S, which is a common substitution found in HIV-1 subtype-C [
34].
3.6.3. Core Domain
The core domain spans from positions 38 to 48 and is involved in interacting with CYCT1 for transactivation and mediating cofactor binding[
32,
35]. An important amino acid motif is a serine (S) residue at position 46 that is phosphorylated by protein kinase (PKR), which is activated by RNA for efficient of LTR transactivation and viral replication. This serine residue was conserved in most (92%) of our patients’ Tat sequences. Patient AK08 had a substitution of S46Y and patient AK68 had S46P substitution in 2 sequences. There were three highly variable amino acids in the core domain of our patients’ Tat sequences at M39, T40, and A42, with 12%, 45% and 13% conservation respectively. However, these amino acids have not been found to be critical for Tat function. The M39 residue was conserved in only 37 sequences in patients AK68, AK78 and AK92, while other patients’ sequences had substitutions with M39L, M39I or with a conservative amino acid M39T. Another amino acid position, A42, was conserved only in 39 Tat sequences, mostly substituted with glycine, a conservative amino acid, likely not affecting Tat functions.
3.6.4. Arginine-Rich Domain
The arginine-rich domain contains a highly conserved region critical for nuclear localization from positions 49 to 57 (
49RKKRRQRRR
57), which facilitates export of Tat to the nucleus to interact with TAR and enhance viral transcription [
30]. Deletion of the arginine-rich domain results in inhibition of HIV-1 LTR transactivation and accumulation of Tat protein in the cytoplasm of the cell[
36]. The arginine-rich region also plays a role in the secretion and internalization of Tat protein into neighboring cells through interaction with surface proteins[
17]. The arginine-rich domain was highly conserved in our patients’ sequences, with conservation at each amino acid residue in our patients’ sequences ranging from 79-100%. Variants at R57 has been shown to reduce uptake of Tat in uninfected cells that alters the production of inflammatory cytokines [
37]. In our patients’ sequences, 93% showed conservation of R57. The most common substitution was with a conservative amino acid, R57K (6%). A substitution of R57Q was found in some sequences of patients AK36, AK83, and AK100. In patients AK36, AK68 and AK99, there was variation at position R53 and Q54 with the following substitutions: R53K (conservative), Q54H and Q54P. Additionally, two important lysine(K) residues are found in the arginine-rich domain that serve as acetylation sites for nuclear localization[
38]. The lysine at 50 was conserved in 297 (100%) of sequences and at position 51 in 296 sequences. An addition of a glycine residue was found at the end of the arginine-rich domain in patients AK19, AK92, and AK99. The arginine-rich domain which is necessary for Tat localization, secretion and uptake were conserved in our 20 HIV
+ patients’ Tat sequences.
3.6.5. Glutamine-Rich Domain
The glutamine-rich domain contains amino acid residues spanning positions 59-72 of Tat protein, which has glutamine residues at position 67 and 72 that were fully conserved in our patients’ sequences. Serine (S) and threonine (T) residues at position 62, 64, and 68 are phosphorylated by PKR and mutations in these amino acid residues lead to decreased phosphorylation and HIV-1 LTR transactivation [
39]. However, even in studies with all three positions, 62, 64, and 68 mutated, transactivation still occurs, but with a lesser efficiency [
39]. Additionally, substitution of at S68 show the greatest transactivation deficits [
39]. In our 20 HIV
+ patients’ Tat sequences, the S at position 62 was conserved in most 85% of sequences. Substitutions at this position included S62R, S62G, S62N and S62D. The T at position 64 was conserved in 172 out of 297 (58%) sequences and common substitutions were T64N, T64S (conservative), T64A, T64I and T64D. Finally, the S at position 68 was conserved in 180 out of 297 (61%) sequences and the substitutions were S68P, S68N and S68D. Overall, this domain was conserved in most patients’ sequences, whereas some patients’ sequences had substitutions that may decrease the function of this domain.
3.7. CTL Epitopes in Tat Regions Sequences from HIV-1-Infected Older Patients on Long Term ART
CTL responses through MHC-1 pathway are induced during primary infection in HIV-1-infected individuals to control viral replication by targeting viral protein fragments. Regulatory proteins such as Tat and Rev of HIV-1 are targeted by cytotoxic T lymphocytes in HIV-1 infected individuals [
40]. Mutations in HIV-1 Tat may allow the virus to escape control of infection by CTL responses. Therefore, we analyzed the previously identified CTL epitopes in Tat deduced amino acid sequences from our 20 HIV
+ patients on long-term ART.
Multiple studies have identified a possible epitope located between amino acid positions 2-11 in the Tat with the sequence EPVDPRLEPW recognized by HLA-B52 [
40,
41], HLA-B53 and HLA-B58 in subtype B [
42]. This epitope was conserved in 4 out of our 20 patients’ Tat sequences, while variations occurred in the other 16 patients’ sequences. The most common variant was a substitution at the arginine (R) at position 7, where only 58% of sequences were found to be conserved. Most common variants in this region are R7S, R7K, R7E and R7N.
Another CTL epitope has been identified with the sequence QPKTACTNCY located in the Tat protein from position 17-26 [
41]. This CTL epitope was fully conserved in 2 out of 20 patients’ sequences, whereas there was a considerable variation in most patients’ sequences. The asparagine (N) residue at position 24 had the lowest conservation where only 37% of sequences retained the asparagine residue with the most common substitutions being N24T, N24K, N24A, or N24Y in 14 patients’ sequences.
Another CTL epitope identified is located at amino acid positions 24-32 in the Tat with the sequence NCYCKKCCF[
43]. This CTL epitope was conserved in one of our HIV
+ patient’s sequences. A common substitution occurred in 14 patients’ sequences at position 24, where N was substituted with N24T, N24K, N24A, or N24Y. In position 29, 10 patients had a substitution with K29R, K29Q, or K29N. In position 32, 12 patients had substitutions with F32Y, F32L, F32M, F32S or F32W.
There were some additional epitopes identified, including from positions 30-37, CCFHCQVC, positions 39-49, ITKGLGISYGR in subtype B, and positions 38-47, FQTKGLGISY, in subtype C, [
42] which was found be generally variable in our patients’ Tat sequences. The high variability within the regions of CTL epitopes suggest that these variants have escaped the host’s immune system. In these patients on long term ART with controlled viremia, variation in the tat gene CTL epitopes suggest persistence of CTL escape mutants.
4. Discussion
Since Tat is essential for HIV-1 replication and pathogenesis [
13,
14,
44], and continues to be secreted in patients on ART with undetectable viral loads [
20], it might play an important role in viral persistence and survival in ART-mediated, virally suppressed HIV-infected patients’ reservoir cells. Our study characterized the molecular, biological, and immunological properties of 297
tat gene sequences from 20 HIV
+ older patients on long term ART who have mostly achieved undetectable viral loads and improved CD4 T cell counts. Our HIV
+ patients’
tat sequences showed a low degree of genetic variability and a high frequency of intact open reading frames with mostly conserved functional domains. This suggests the importance of Tat protein in HIV persistence and survival in HIV-infected patients with controlled viremia on ART, which are consistent with previous studies that analyzed
tat sequences in HIV-infected individuals[
44,
45,
46]. In addition, the previously characterized CTL epitopes were found to be variable in our HIV
+ patients’ sequences, suggesting that these quasispecies escaped CTL response and established persistence in these infected patients.
Naturally occurring mutations in Tat may modulate transactivation or other functions and impact the establishment and reversal of HIV-1 latency and reactivation. Functional domains in Tat critical for its activity, including TAR binding, transactivation and nuclear localization were highly conserved in our older patients’ deduced amino acid sequences of Tat, consistent with earlier studies on HIV-infected patients on ART[
45]. The first functional domain of Tat consists of the proline-rich amino-terminal, cysteine-rich and core regions [
29,
47,
48] which is essential for TAR transactivation, viral transcription and replication[
29,
46]. In our patients’ Tat sequences, the important motifs in the proline-rich, cysteine-rich, and core regions were mostly conserved, suggesting that Tat from these patients’ sequences have most likely maintained the ability to bind to TAR and influence viral transcription and replication necessary for viral survival and persistence despite viral suppression by ART. The second functional domain consists of the basic and glutamine rich regions that are responsible for the other functions of Tat such as nuclear localization, cellular uptake and binding of Tat to TAR RNA [
30]. Our patients’ Tat sequences showed conservation in both basic and glutamine regions like shown other studies[
44,
45] but in the glutamine-rich region, some of the important amino acids such as T64 and S68 had substitutions that may have some reduced functions of Tat [
39], that may be sufficient for a low level of viral transcription needed for viral persistence in these patients on ART. Earlier studies that we have conducted characterized the HIV-1 envelope V3 region and
vpr gene from mostly the same HIV
+ patients which showed a conservation of functional domains [
49,
50], corroborating with the current data on
tat that suggests that HIV- genomes in these patients’ are generally functional.
Although HIV typically produces a high level of genetic diversity due to error prone reverse transcriptase [
6], our current study showed that the
tat sequences from HIV
+ patients exhibit a low degree of genetic variability and lower estimates of genetic diversity most likely due to suppression of viral replication by ART. Since the virus is unable to replicate, mutations from errors by reverse transcriptase do not accumulate leading to lower measures of genetic diversity. Our previous studies on the V3 region and
vpr gene sequences from mostly the same patients on long term ART showed a low degree of genetic variability and estimates of genetic diversity [
49,
50]. In addition, continuous CD8 T cell responses to low levels of HIV antigen in our HIV
+ patients on ART may also be responsible for the low level of genetic diversity in these patients because CD8
+ T cells have been implicated to control of virus replication in infected cells where HIV persists under ART [
51,
52]. Of note, previously characterized CTL epitopes in Tat sequences were found to be generally variable in our patients’ Tat sequences, suggesting that the HIV proviral DNA persisting in HIV
+ patients on ART carry CTL escape mutants that were probably generated before the initiation of ART[
53,
54]. Since there is low viral replication with successful use of ART, not enough newer viral proteins are not being made and loaded onto MHC I to activate naïve CD8 T cells to generate new CTL against these epitopes which would eliminate these latently infected cells [
4,
55].
Since Tat is essential for viral replication and pathogenesis, targeting Tat for therapeutic and curative strategies may provide significant improvement in outcomes over the current HIV regimens. Exogenous Tat crosses the blood brain barrier causing neurotoxic effects on the nervous system resulting in HAND among HIV+ patients [
56,
57]. Of interest, the C31S substitution in Tat in HIV-1 subtype C, and a few of our sequences, induces an activated phenotype in endothelial cells, enhancing migration, invasion, and in vitro morphogenesis potential [
58]. The C31S variant leads to decreased chemotactic function while maintaining transactivation properties, which may alter neurotoxic effects of Tat in some patients [
34]. Presence of higher levels of Tat antibodies in HIV-1 infected patients have been shown to delay HIV disease progression and reduce likelihood of HAND [
57]. Therefore, Tat is a protein of interest in reducing neuropathies associated with HIV-1 infection. The genetic characterization of 297 Tat sequences from 20 HIV+ patients with variability in several domains may aid in the identification of a variant that may be more immunogenic.
Limitations of this study are that the HIV+ cohort included only 20 patients. Future studies should be conducted to further characterize the tat gene in more HIV older patients, including biological characterization of several of these unique sequences by creating chimeric HIV-1. The data presented in this study on the characterization of HIV-1 tat genes in HIV-infected patients on long term ART have provided new insights in viral persistence and survival in these patients and may aid in the development of strategies to eliminate HIV from the infected population.
Author Contributions
Conceptualization, N.A.; methodology, N.A, N.K., N.M.; validation, N.A., N.K., N.M.; formal analysis, N.A., N.K., N.M.; investigation, N.A., N.K., N.M., A.D., N.C.; resources, N.A., N.K., N.M, SK.; data curation, N.K., N.M.; writing—original draft preparation, N.A., N.K.; writing—review and editing, N.A., N.K. S.K; visualization, N.A., N.K.; supervision, N.A., S.K. ; project administration, N.A., N.K., N.M.; funding acquisition, N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a small internal funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of the University of Arizona in Tucson, AZ (IRB Approval# 1412586476).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The nucleotide sequences with accession numbers OR265643 to OR265939 were submitted to GenBank. We confirm that the data related to the study are available within the article.
Acknowledgments
The authors thank all patients for their participation in this study. The authors also thank David Harris for PBMC isolation and storage at the Biorepository and Maria Love and Luiza Rodrigues-Samora for assistance in initiation of this work.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
-
HIV Surveillance Report, 2019; Centers for Disease Control and Prevention: 2021.
- May, M.T.; Gompels, M.; Delpech, V.; Porter, K.; Orkin, C.; Kegg, S.; Hay, P.; Johnson, M.; Palfreeman, A.; Gilson, R.; et al. Impact on life expectancy of HIV-1 positive individuals of CD4+ cell count and viral load response to antiretroviral therapy. AIDS 2014, 28, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.L.; Chao, C.R.; Leyden, W.A.; Xu, L.; Quesenberry, C.P., Jr.; Klein, D.B.; Towner, W.J.; Horberg, M.A.; Silverberg, M.J. Narrowing the Gap in Life Expectancy Between HIV-Infected and HIV-Uninfected Individuals With Access to Care. J Acquir Immune Defic Syndr 2016, 73, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Behrens, N.E.; Wertheimer, A.; Klotz, S.A.; Ahmad, N. Reduction in terminally differentiated T cells in virologically controlled HIV-infected aging patients on long-term antiretroviral therapy. PLoS One 2018, 13, e0199101. [Google Scholar] [CrossRef]
- Behrens, N.E.; Wertheimer, A.; Love, M.B.; Klotz, S.A.; Ahmad, N. Evaluation of HIV-specific T-cell responses in HIV-infected older patients with controlled viremia on long-term antiretroviral therapy. PLoS One 2020, 15, e0236320. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.D.; Bebenek, K.; Kunkel, T.A. The accuracy of reverse transcriptase from HIV-1. Science 1988, 242, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Pertea, M.; Rongvaux, A.; Wang, L.; Durand, C.M.; Ghiaur, G.; Lai, J.; McHugh, H.L.; Hao, H.; Zhang, H.; et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015, 517, 381–385. [Google Scholar] [CrossRef]
- Chun, T.W.; Engel, D.; Berrey, M.M.; Shea, T.; Corey, L.; Fauci, A.S. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 1998, 95, 8869–8873. [Google Scholar] [CrossRef]
- Chun, T.W.; Carruth, L.; Finzi, D.; Shen, X.; DiGiuseppe, J.A.; Taylor, H.; Hermankova, M.; Chadwick, K.; Margolick, J.; Quinn, T.C.; et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997, 387, 183–188. [Google Scholar] [CrossRef]
- Finzi, D.; Blankson, J.; Siliciano, J.D.; Margolick, J.B.; Chadwick, K.; Pierson, T.; Smith, K.; Lisziewicz, J.; Lori, F.; Flexner, C.; et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999, 5, 512–517. [Google Scholar] [CrossRef]
- Clark, E.; Nava, B.; Caputi, M. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 2017, 8, 27569–27581. [Google Scholar] [CrossRef]
- Berkhout, B.; Silverman, R.H.; Jeang, K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, M.B.; Baltimore, D.; Frankel, A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A 1991, 88, 4045–4049. [Google Scholar] [CrossRef] [PubMed]
- Faust, T.B.; Binning, J.M.; Gross, J.D.; Frankel, A.D. Making Sense of Multifunctional Proteins: Human Immunodeficiency Virus Type 1 Accessory and Regulatory Proteins and Connections to Transcription. Annu Rev Virol 2017, 4, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Izmailova, E.; Bertley, F.M.; Huang, Q.; Makori, N.; Miller, C.J.; Young, R.A.; Aldovini, A. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat Med 2003, 9, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Secchiero, P.; Zella, D.; Capitani, S.; Gallo, R.C.; Zauli, G. Extracellular HIV-1 tat protein up-regulates the expression of surface CXC-chemokine receptor 4 in resting CD4+ T cells. J Immunol 1999, 162, 2427–2431. [Google Scholar] [CrossRef]
- Ensoli, B.; Buonaguro, L.; Barillari, G.; Fiorelli, V.; Gendelman, R.; Morgan, R.A.; Wingfield, P.; Gallo, R.C. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol 1993, 67, 277–287. [Google Scholar] [CrossRef]
- Carvallo, L.; Lopez, L.; Fajardo, J.E.; Jaureguiberry-Bravo, M.; Fiser, A.; Berman, J.W. HIV-Tat regulates macrophage gene expression in the context of neuroAIDS. PLoS One 2017, 12, e0179882. [Google Scholar] [CrossRef]
- Jiang, Y.; Chai, L.; Fasae, M.B.; Bai, Y. The role of HIV Tat protein in HIV-related cardiovascular diseases. J Transl Med 2018, 16, 121. [Google Scholar] [CrossRef]
- Mediouni, S.; Darque, A.; Baillat, G.; Ravaux, I.; Dhiver, C.; Tissot-Dupont, H.; Mokhtari, M.; Moreau, H.; Tamalet, C.; Brunet, C.; et al. Antiretroviral therapy does not block the secretion of the human immunodeficiency virus tat protein. Infect Disord Drug Targets 2012, 12, 81–86. [Google Scholar] [CrossRef]
- Thomas H. Jukes, C.R.C. Mammilian Protein Metabolism; Munro, H.N., Ed.; Academic Press: 1969.
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor Popul Biol 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Nei, M. Molecular evolutionary genetics; New York: Columbia University Press: 1987.
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Rayne, F.; Debaisieux, S.; Yezid, H.; Lin, Y.L.; Mettling, C.; Konate, K.; Chazal, N.; Arold, S.T.; Pugniere, M.; Sanchez, F.; et al. Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J 2010, 29, 1348–1362. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; de, A.Z.P.M. Genetic drift of human immunodeficiency virus type 1? J Virol 1998, 72, 886–887. [Google Scholar] [CrossRef] [PubMed]
- Teto, G.; Fonsah, J.Y.; Tagny, C.T.; Mbanya, D.; Nchindap, E.; Kenmogne, L.; Fokam, J.; Njamnshi, D.M.; Kouanfack, C.; Njamnshi, A.K.; et al. Molecular and Genetic Characterization of HIV-1 Tat Exon-1 Gene from Cameroon Shows Conserved Tat HLA-Binding Epitopes: Functional Implications. Viruses 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Kuppuswamy, M.; Subramanian, T.; Srinivasan, A.; Chinnadurai, G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res 1989, 17, 3551–3561. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.A.; Harrich, D.; Pearson, L.; Mitsuyasu, R.; Gaynor, R.B. Functional domains required for tat-induced transcriptional activation of the HIV-1 long terminal repeat. EMBO J 1988, 7, 3143–3147. [Google Scholar] [CrossRef]
- Hauber, J.; Malim, M.H.; Cullen, B.R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 1989, 63, 1181–1187. [Google Scholar] [CrossRef]
- Rappaport, J.; Lee, S.J.; Khalili, K.; Wong-Staal, F. The acidic amino-terminal region of the HIV-1 Tat protein constitutes an essential activating domain. New Biol 1989, 1, 101–110. [Google Scholar]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Pierleoni, R.; Menotta, M.; Antonelli, A.; Sfara, C.; Serafini, G.; Dominici, S.; Laguardia, M.E.; Salis, A.; Damonte, G.; Banci, L.; et al. Effect of the redox state on HIV-1 tat protein multimerization and cell internalization and trafficking. Mol Cell Biochem 2010, 345, 105–118. [Google Scholar] [CrossRef]
- Ranga, U.; Shankarappa, R.; Siddappa, N.B.; Ramakrishna, L.; Nagendran, R.; Mahalingam, M.; Mahadevan, A.; Jayasuryan, N.; Satishchandra, P.; Shankar, S.K.; et al. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol 2004, 78, 2586–2590. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.E.; Wei, P.; KewalRamani, V.N.; Mayall, T.P.; Herrmann, C.H.; Rice, A.P.; Littman, D.R.; Jones, K.A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 1998, 12, 3512–3527. [Google Scholar] [CrossRef] [PubMed]
- Orsini, M.J.; Debouck, C.M. Inhibition of human immunodeficiency virus type 1 and type 2 Tat function by transdominant Tat protein localized to both the nucleus and cytoplasm. J Virol 1996, 70, 8055–8063. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.P.; Ajasin, D.O.; Ramasamy, S.; DesMarais, V.; Eugenin, E.A.; Prasad, V.R. A Naturally Occurring Polymorphism in the HIV-1 Tat Basic Domain Inhibits Uptake by Bystander Cells and Leads to Reduced Neuroinflammation. Scientific Reports 2019, 9, 3308. [Google Scholar] [CrossRef]
- He, M.; Zhang, L.; Wang, X.; Huo, L.; Sun, L.; Feng, C.; Jing, X.; Du, D.; Liang, H.; Liu, M.; et al. Systematic Analysis of the Functions of Lysine Acetylation in the Regulation of Tat Activity. PLoS One 2013, 8, e67186. [Google Scholar] [CrossRef]
- Endo-Munoz, L.; Warby, T.; Harrich, D.; McMillan, N.A. Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J 2005, 2, 17. [Google Scholar] [CrossRef]
- Addo, M.M.; Altfeld, M.; Rosenberg, E.S.; Eldridge, R.L.; Philips, M.N.; Habeeb, K.; Khatri, A.; Brander, C.; Robbins, G.K.; Mazzara, G.P.; et al. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci U S A 2001, 98, 1781–1786. [Google Scholar] [CrossRef]
- Yusim, K.; Kesmir, C.; Gaschen, B.; Addo, M.M.; Altfeld, M.; Brunak, S.; Chigaev, A.; Detours, V.; Korber, B.T. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol 2002, 76, 8757–8768. [Google Scholar] [CrossRef]
- Chakraborty, S.; Rahman, T.; Chakravorty, R. Characterization of the Protective HIV-1 CTL Epitopes and the Corresponding HLA Class I Alleles: A Step towards Designing CTL Based HIV-1 Vaccine. Adv Virol 2014, 2014, 321974. [Google Scholar] [CrossRef]
- Jones, N.A.; Wei, X.; Flower, D.R.; Wong, M.; Michor, F.; Saag, M.S.; Hahn, B.H.; Nowak, M.A.; Shaw, G.M.; Borrow, P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med 2004, 200, 1243–1256. [Google Scholar] [CrossRef]
- Husain, M.; Hahn, T.; Yedavalli, V.R.; Ahmad, N. Characterization of HIV type 1 tat sequences associated with perinatal transmission. AIDS Res Hum Retroviruses 2001, 17, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Ronsard, L.; Lata, S.; Singh, J.; Ramachandran, V.G.; Das, S.; Banerjea, A.C. Molecular and Genetic Characterization of Natural HIV-1 Tat Exon-1 Variants from North India and Their Functional Implications. PLOS ONE 2014, 9, e85452. [Google Scholar] [CrossRef] [PubMed]
- Spector, C.; Mele, A.R.; Wigdahl, B.; Nonnemacher, M.R. Genetic variation and function of the HIV-1 Tat protein. Med Microbiol Immunol 2019, 208, 131–169. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, K.; Fujinaga, K.; Okamoto, T. HIV Tat/P-TEFb Interaction: A Potential Target for Novel Anti-HIV Therapies. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Selby, M.J.; Peterlin, B.M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell 1990, 62, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Behrens, N.E.; Love, M.; Bandlamuri, M.; Bernhardt, D.; Wertheimer, A.; Klotz, S.A.; Ahmad, N. Characterization of HIV-1 Envelope V3 Region Sequences from Virologically Controlled HIV-Infected Older Patients on Long Term Antiretroviral Therapy. AIDS Res Hum Retroviruses 2021, 37, 233–245. [Google Scholar] [CrossRef]
- Love, M.; Samora, L.; Barker, D.; Zukosky, P.; Kummet, N.; Ahmad, A.; Bernhardt, D.; Tripathi, M.; Klotz, S.; Ahmad, N. Genetic Analysis of HIV-1 vpr Sequences from HIV-Infected Older Patients on Long-Term Antiretroviral Therapy. Curr HIV Res 2022, 20, 309–320. [Google Scholar] [CrossRef]
- McBrien, J.B.; Kumar, N.A.; Silvestri, G. Mechanisms of CD8(+) T cell-mediated suppression of HIV/SIV replication. Eur J Immunol 2018, 48, 898–914. [Google Scholar] [CrossRef]
- Dampier, W.; Nonnemacher, M.R.; Mell, J.; Earl, J.; Ehrlich, G.D.; Pirrone, V.; Aiamkitsumrit, B.; Zhong, W.; Kercher, K.; Passic, S.; et al. HIV-1 Genetic Variation Resulting in the Development of New Quasispecies Continues to Be Encountered in the Peripheral Blood of Well-Suppressed Patients. PLOS ONE 2016, 11, e0155382. [Google Scholar] [CrossRef]
- Kearney, M.F.; Spindler, J.; Shao, W.; Yu, S.; Anderson, E.M.; O'Shea, A.; Rehm, C.; Poethke, C.; Kovacs, N.; Mellors, J.W.; et al. Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy. PLOS Pathogens 2014, 10, e1004010. [Google Scholar] [CrossRef]
- Bozzi, G.; Simonetti, F.R.; Watters, S.A.; Anderson, E.M.; Gouzoulis, M.; Kearney, M.F.; Rote, P.; Lange, C.; Shao, W.; Gorelick, R.; et al. No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: Implications for HIV eradication. Science Advances 2019, 5, eaav2045. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.A.; Clutton, G.; Goonetilleke, N. Harnessing CD8(+) T Cells Under HIV Antiretroviral Therapy. Front Immunol 2019, 10, 291. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Hou, J.; Su, B.; Jiang, T.; Guo, C.; Wang, W.; Zhang, Y.; Chang, B.; Wu, H.; Zhang, T. The Prevalence of Frascati-Criteria-Based HIV-Associated Neurocognitive Disorder (HAND) in HIV-Infected Adults: A Systematic Review and Meta-Analysis. Front Neurol 2020, 11, 581346. [Google Scholar] [CrossRef] [PubMed]
- Bachani, M.; Sacktor, N.; McArthur, J.C.; Nath, A.; Rumbaugh, J. Detection of anti-tat antibodies in CSF of individuals with HIV-associated neurocognitive disorders. J Neurovirol 2013, 19, 82–88. [Google Scholar] [CrossRef]
- Menon, M.; Budhwar, R.; Shukla, R.N.; Bankar, K.; Vasudevan, M.; Ranga, U. The Signature Amino Acid Residue Serine 31 of HIV-1C Tat Potentiates an Activated Phenotype in Endothelial Cells. Front Immunol 2020, 11, 529614. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).