Submitted:
27 September 2023
Posted:
28 September 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
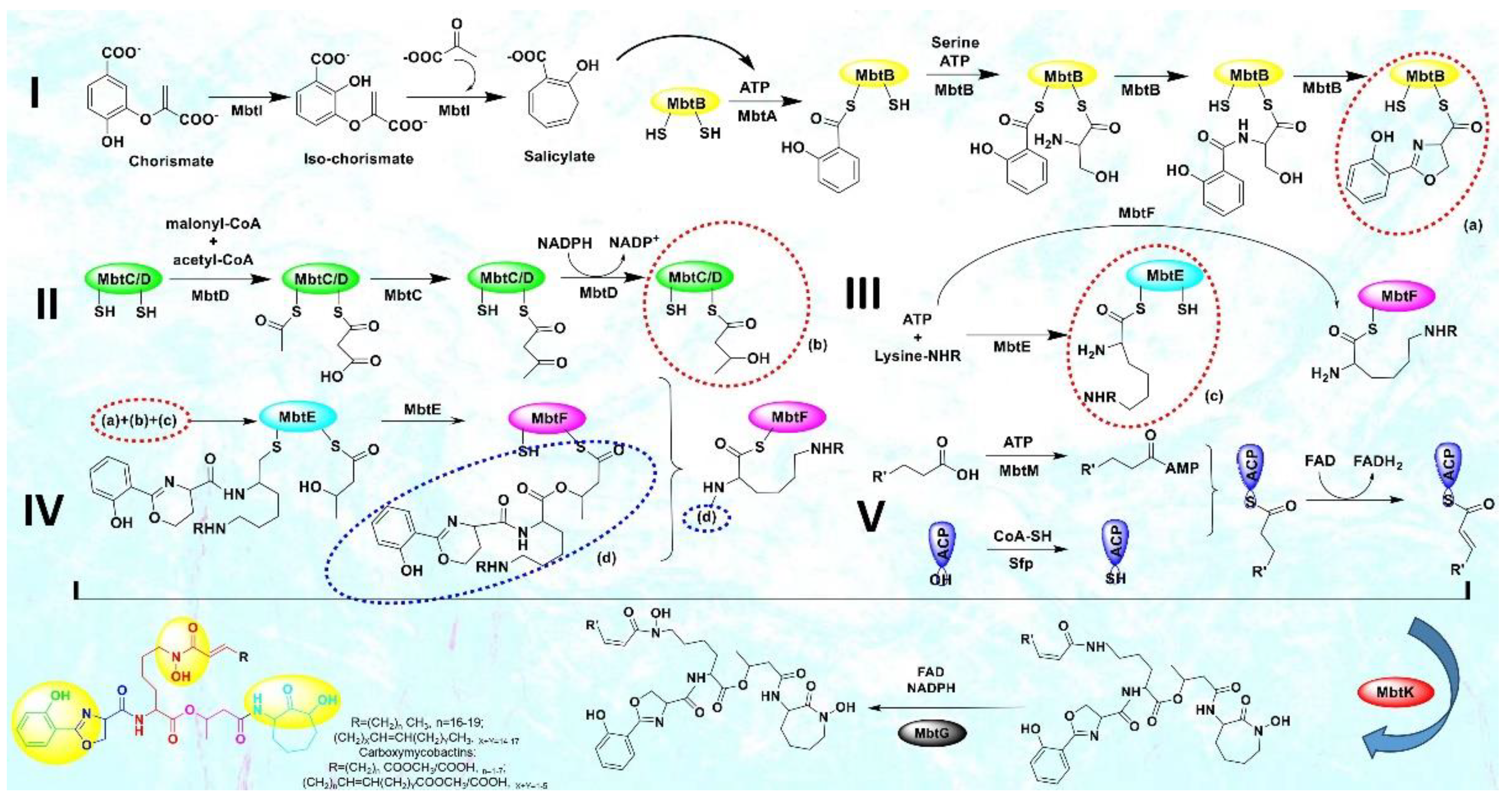
2. Materials and Methods
2.1. System Specifications and Software Employed
2.2. Ligand Preparation
2.3. Protein Preparation
2.4. Identification of Binding Site and Receptor Grid Generation
2.5. Docking-Based Virtual Screening Studies
2.6. Molecular Dynamics Studies
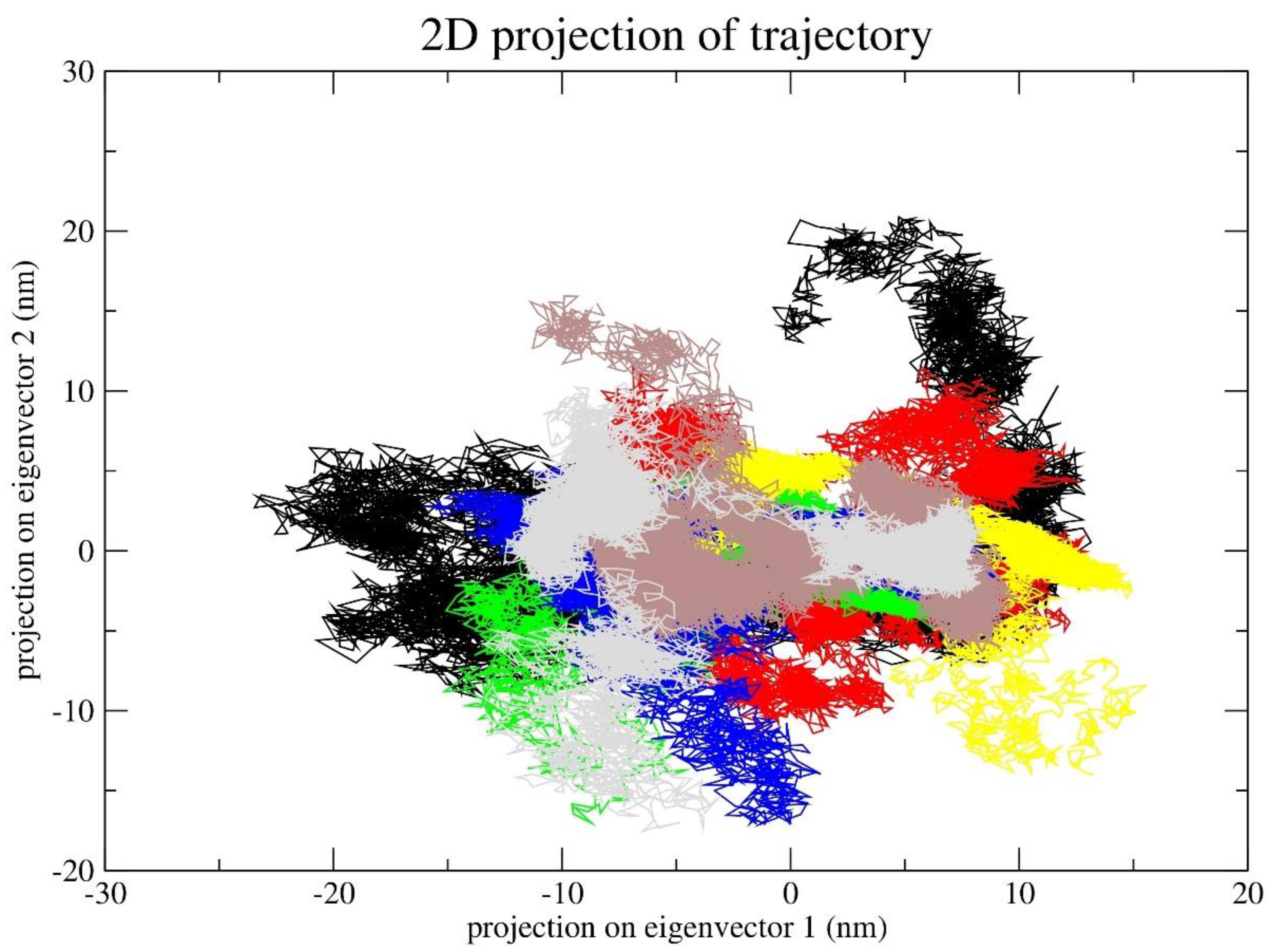
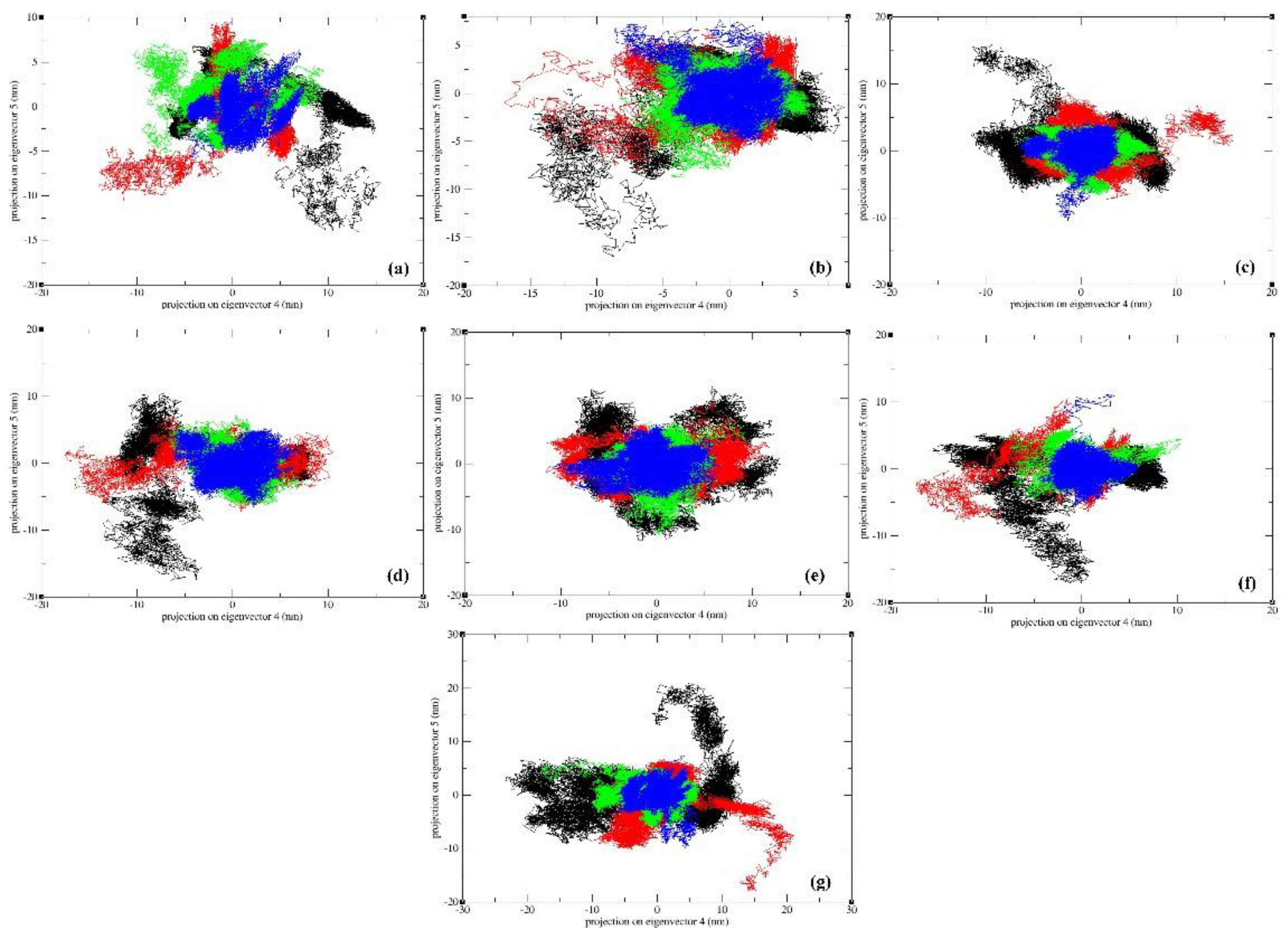
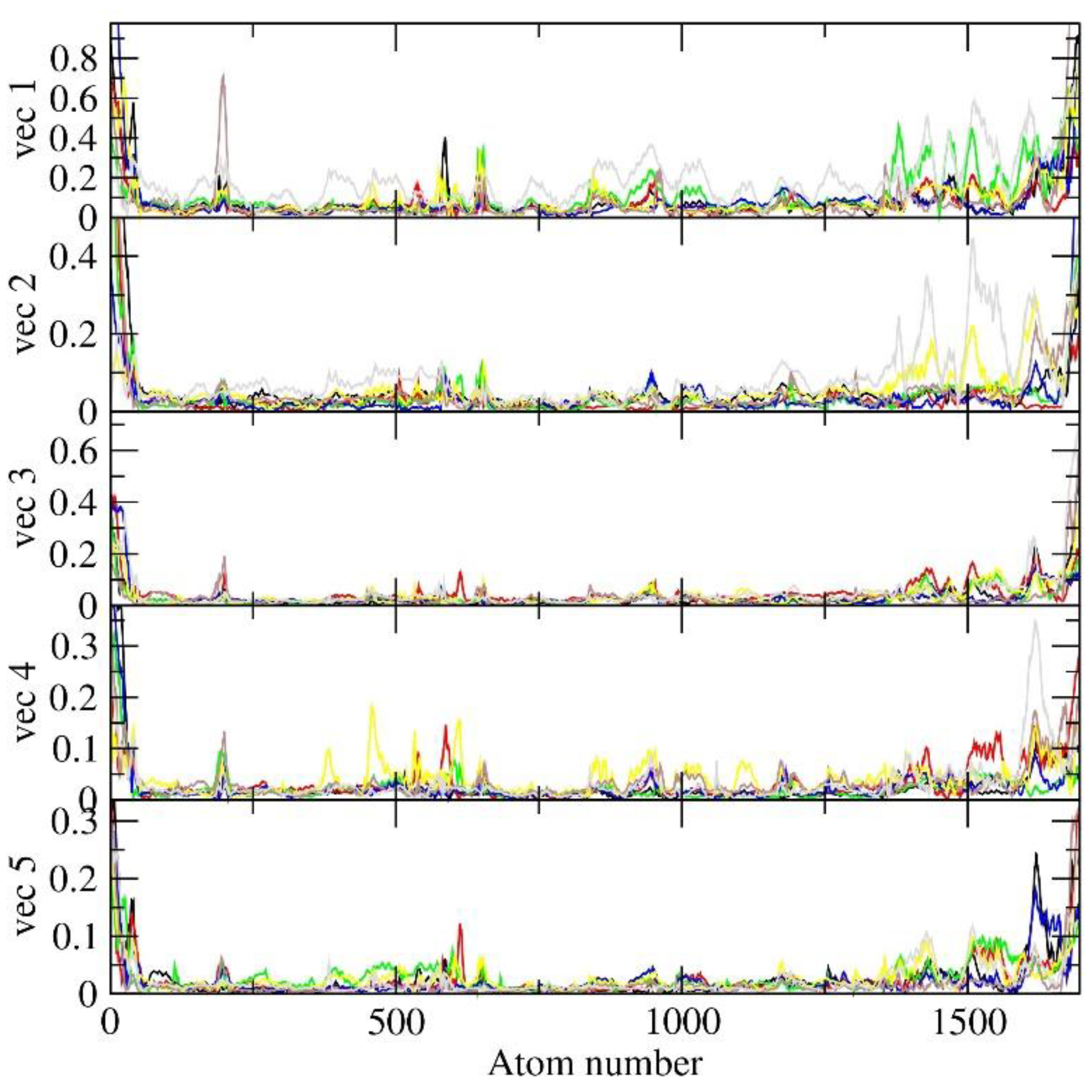
2.7. Principal Component Analysis (PCA)
3. Results
3.1. Molecular Docking Studies on MbtA
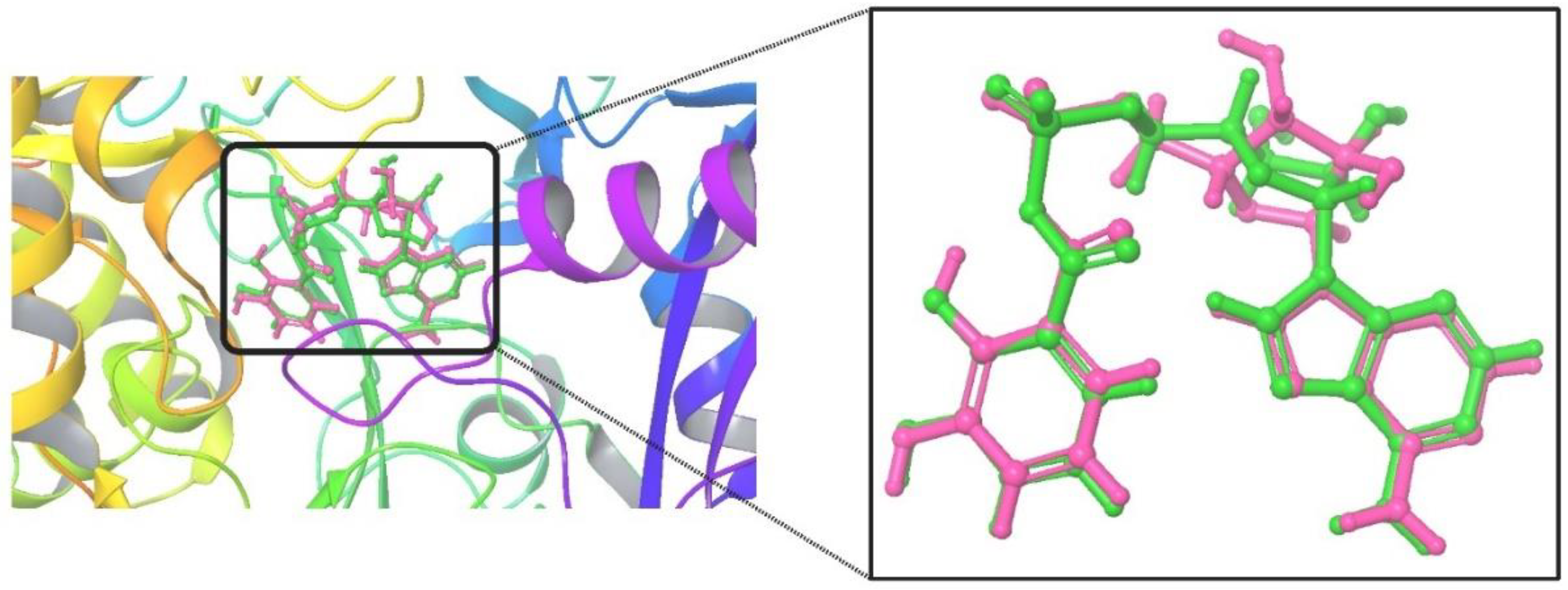
3.1.1. Validation of Docking Procedure
3.1.2. Virtual Screening of FDA-Reported Library Through Molecular Docking
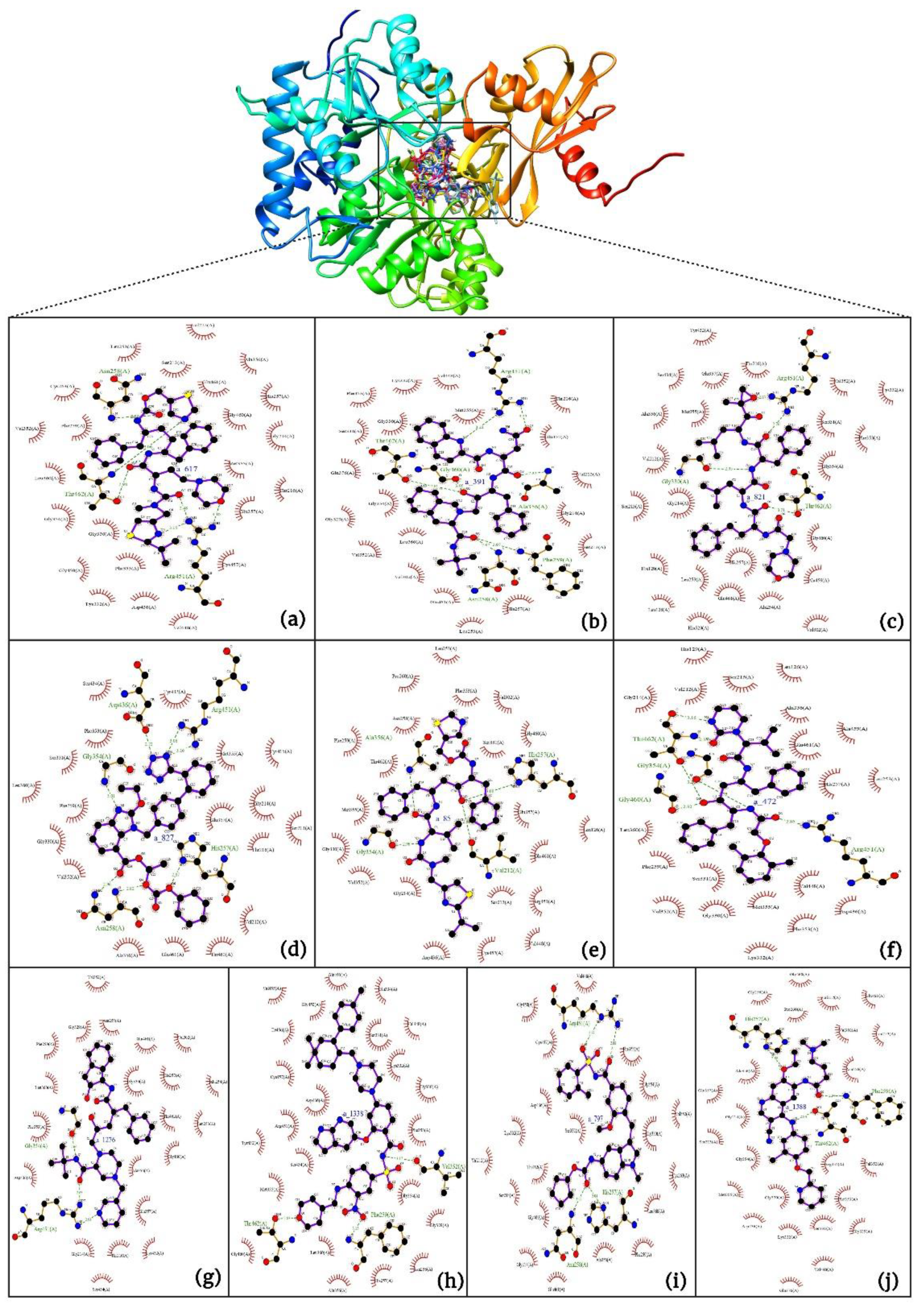
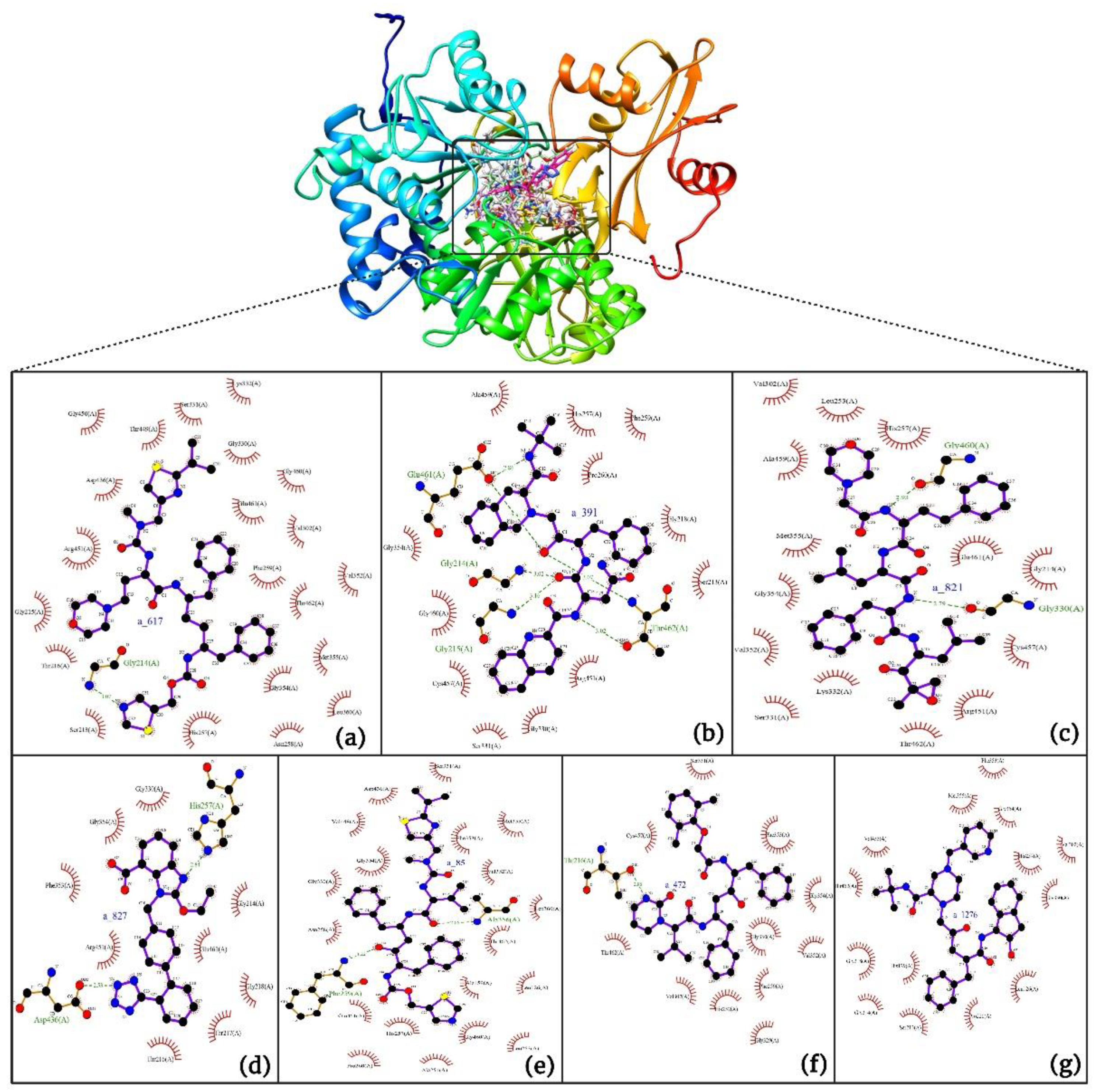
- a_617: Cobicistat – MbtA complex (Figure 6a)
- 2.
- a_391: Saquinavir – MbtA complex (Figure 6b)
- 3.
- a_821: Carfilzomib – MbtA complex (Figure 6c)
- 4.
- a_827: Candesartan – MbtA complex (Figure 6d)
- 5.
- a_85: Ritonavir – MbtA complex (Figure 6e)
- 6.
- a_472: Lopinavir – MbtA complex (Figure 6f)
- 7.
- a_1276: Indinavir – MbtA complex (Figure 6g)
- 8.
- a_1338: Venetoclax – MbtA complex (Figure 6h)
- 9.
- a_797: Zafirlukast (Accolate) – MbtA complex (Figure 6i)
- 10.
- a_1388: Neratinib – MbtA complex (Figure 6j)
| Sl. No. | Code | Docking interactions with active site amino acid residues | H-bond distance (Å) |
|---|---|---|---|
| 1 | a_617 |
H-bond- Asn258, Thr462, & Arg451 Hydrophobic- Val448, Asp436, Lys332, Phe353, Gly450, Gly330, Gly354, Leu360, Val352, Phe259, Cys263, Leu253, Ser213, Glu461, Ala356, Gly460, His257, Gly214, Met355, Thr216, Glu357, & Cys457 |
2.80, [3.06, 2.98], & [3.15, 2.48, 3.10] |
| 2 | a_391 |
H-bond- Asn258, Thr462, Arg451, Gly460, Ala356, & Phe259 Hydrophobic- His257, Leu253, Glu461, Val302, Leu360, Val352, Gly354, Gly329, Gln376, Ser331, Gly330, Phe353, Lys332, Val448, Met355, Thr216, Glu357, Val212, Gly214, & Ser213 |
2.87, 2.81, [3.23, 3.21], 3.05, 2.92, & [2.87, 3.07] |
| 3 | a_821 |
H-bond- Gly330, Thr462, & Arg451 Hydrophobic- Ala254, His257, Glu461, His523, Leu253, Leu126, His129, Gly214, Ser213, Val212, Met355, Ala356, Ser434, Glu357, Tyr432, Thr216, Val352, Lys332, Ser331, Phe353, Gly354, Gly460, & Ala459 |
2.95, 3.21, & 2.76 |
| 4 | a_827 |
H-bond- Asn258, Arg451, Asp436, Gly354, & His257 Hydrophobic-Val212, Thr462, Glu461, Ala356, Val352, Gly330, Phe259, Leu360, Ser331, Phe353, Ser434, Tyr432, Met355, Tyr415, Gly214, Glu357, Thr216, & Ser213 |
[2.82, 2.96], [3.16, 3.06], 2.72, 3.01, 2.87 |
| 5 | a_85 |
H-bond- Ala356, Val212, Gly354, & His257 Hydrophobic- Gly214, Val352, Gly330, Met355, Thr462, Phe259, Asn258, Pro260, Leu253, Phe353, Val302, Ser331, Gly460, Glu357, Leu126, Glu461, Arg451, Ser213, Val448, Cys457, & Asp436 |
2.88, 2.80, 2.96, & 3.08 |
| 6 | a_472 |
H-bond- Gly354, Gly460, Thr462, & Arg451 Hydrophobic- Asp436, Val448, Phe353, Met355, Lys332, Gly330, Ser331, Val352, Phe259, Leu360, Gly214, Val212, His129, Ser213, Leu126, Ala356, Glu461, Ala459, His257, & Leu253 |
2.75, 2.92, 3.16, & 2.86 |
| 7 | a_1276 |
H-bond- Gly354 & Arg451 Hydrophobic- Asp436, Phe353, Leu360, Phe259, Gly329, Val352, Asn258, Glu461, Val302, Gly330, His257, Ala254, Thr462, Leu253, Gly460, Ala356, Glu357, Tyr432, Thr216, Gly214, & Ser434 |
2.66, & [2.63, 2.93] |
| 8 | a_1338 |
H-bond- Val352, Thr462, & Phe259 Hydrophobic- Gly460, Met355, Ser434, Tyr432, Arg451, Asp436, Cys457, Ile456, Gly450, Val455, Glu493, Glu334, Ser331, Val448, Lys332, Gly330, Phe353, Gly354, Gly329, Asn258, His257, Ala356, & Leu360 |
3.17, 2.83, & 3.15 |
| 9 | a_797 |
H-bond- Asn258, His257, & Arg451 Hydrophobic- Glu461, Gly214, Gly460, Ser213, Thr462, Val212, Lys332, Asp436, Ser331, Cys457, Gly450, Val448, Phe353, Gly354, Val352, Gly330, Cys263, Leu360, Phe259, & Ala356 |
3.07, 3.04, & [2.87, 2.88] |
| 10 | a_1388 |
H-bond- His257, Phe259, & Thr462 Hydrophobic- Arg451, Val352, Phe353, Gly329, Ser331, Val448, Gln376, Lys332, Gly330, Asp436, Met355, Gly354, Ser213, Gly214, Glu357, Ala356, Gly256, Pro260, Gly460, Val212, Glu461, Val302, Leu253, & Asn258 |
2.88, 2.94, & 2.94 |
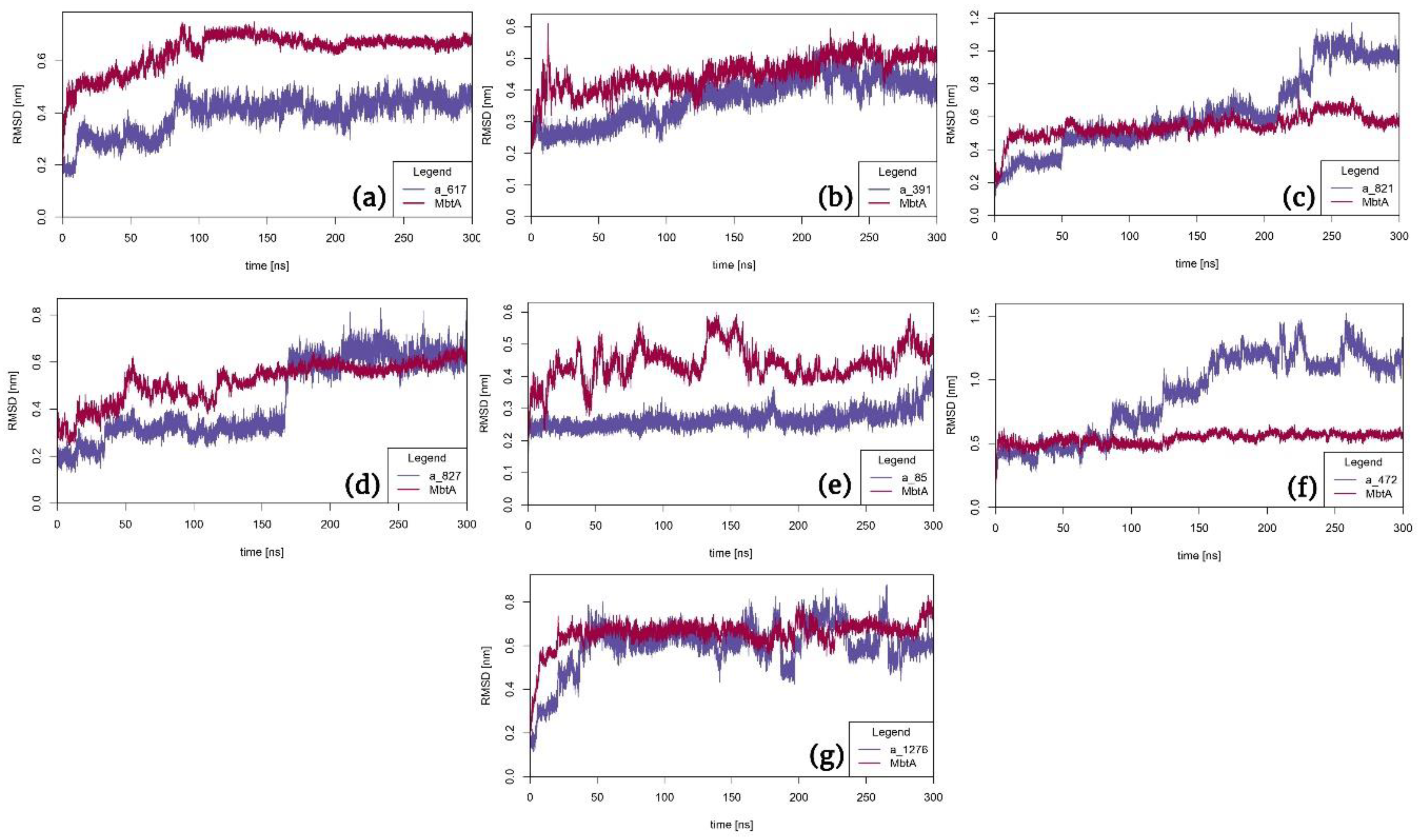
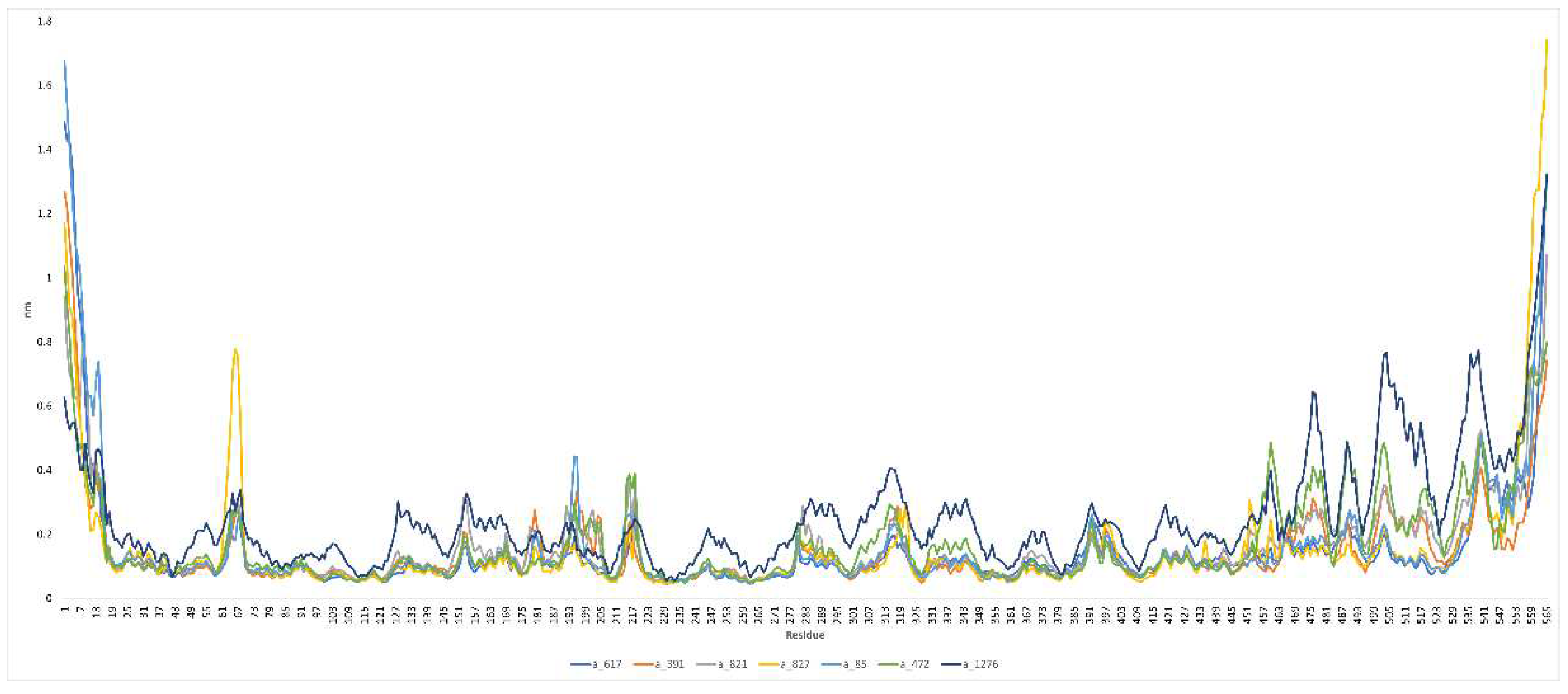
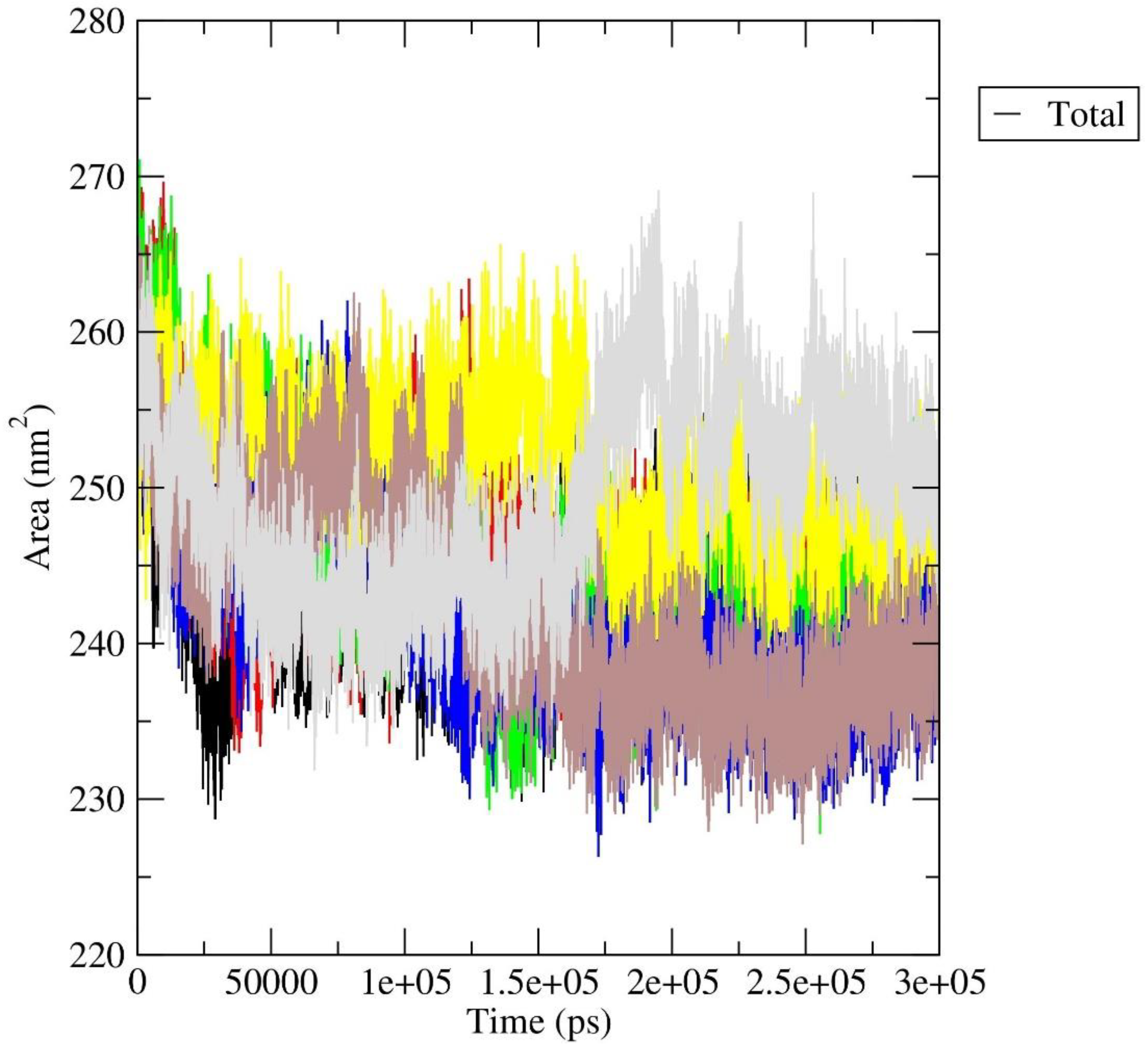
3.2. Molecular Dynamics Simulation Study
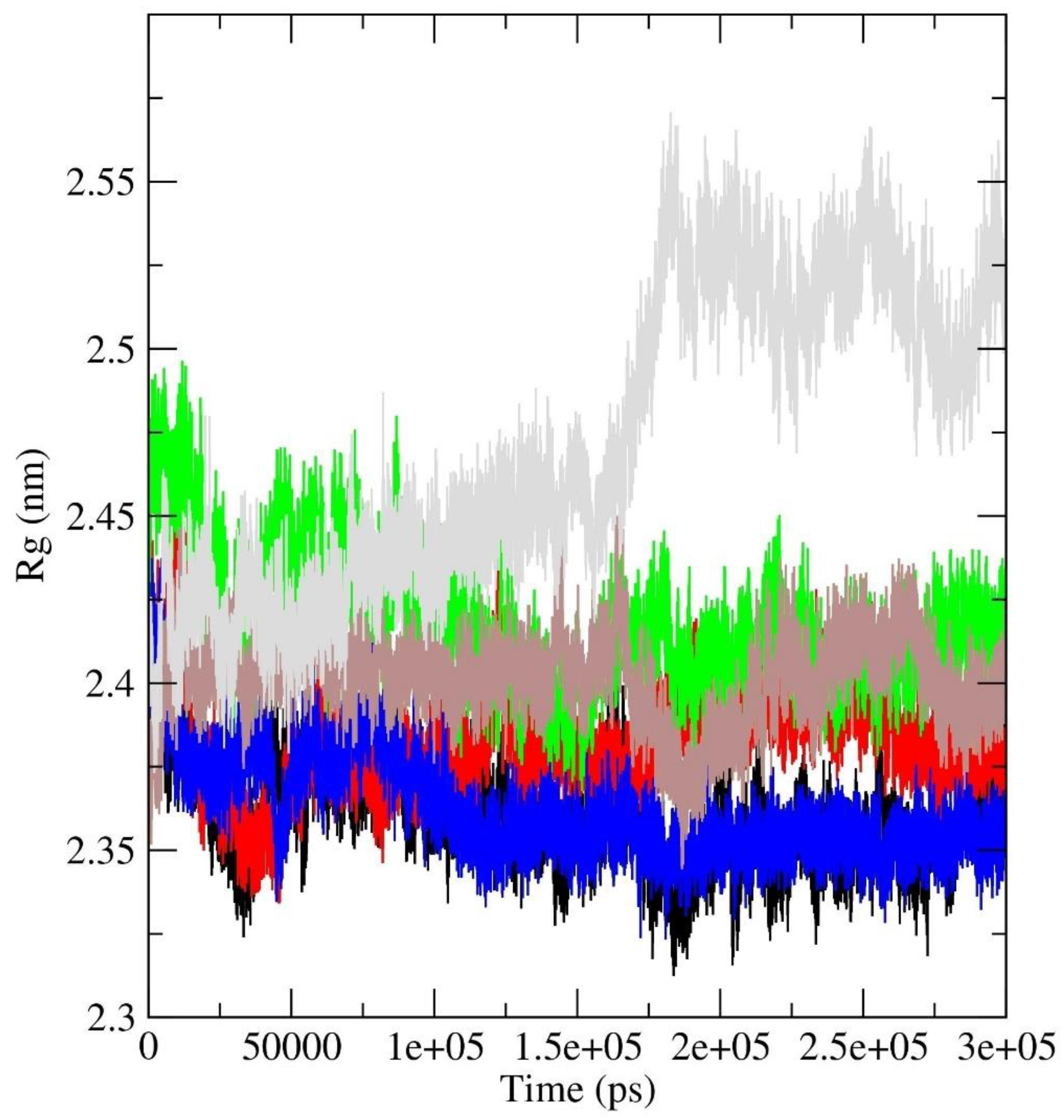
4. Principal Component Analysis (PCA)
5. Discussion
6. Conclusion
7. Future Scope
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Borah, P.; Deb, P.K.; Venugopala, K.N.; Al-Shar’i, N.A.; Singh, V.; Deka, S.; Srivastava, A.; Tiwari, V.; Mailavaram, R.P. Tuberculosis: An Update on Pathophysiology, Molecular Mechanisms of Drug Resistance, Newer Anti-TB Drugs, Treatment Regimens and Host-Directed Therapies. Curr Top Med Chem 2021, 21, 547–570. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Tuberculosis: Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed on 18 September 2023).
- Peddireddy, V.; Doddam, S.N.; Ahmed, N. Mycobacterial Dormancy Systems and Host Responses in Tuberculosis. Front Immunol 2017, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Shyam, M.; Shilkar, D.; Verma, H.; Dev, A.; Sinha, B.N.; Brucoli, F.; Bhakta, S.; Jayaprakash, V. The Mycobactin Biosynthesis Pathway: A Prospective Therapeutic Target in the Battle against Tuberculosis. J Med Chem 2020, 64, 71–100. [Google Scholar] [CrossRef]
- Prasad, R.; Singh, A.; Balasubramanian, V.; Gupta, N. Extensively Drug-Resistant Tuberculosis in India: Current Evidence on Diagnosis & Management. Indian J Med Res 2017, 145, 271. [Google Scholar]
- Velayati, A.A.; Farnia, P.; Hoffner, S. Drug-Resistant Mycobacterium Tuberculosis: Epidemiology and Role of Morphological Alterations. J Glob Antimicrob Resist 2018, 12, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, G.; Jayaprakash, V. Tuberculosis and HIV Responses Threatened by NCOVID-19: A Situation Prompting an in Silico Investigation of Reported MbtA Inhibitors for Combined Inhibition of SARS-CoV-2 and HIV-TB Co-Infection. Struct Chem 2023, 34, 655–679. [Google Scholar] [CrossRef]
- Sibi, D.; Sethi Das, C.; Jibin, V.G.; Silvanose, C.D. Emerging Antibiotic Resistance in Post-COVID-19 Co-Infections. J Clin Med Case Reports 2023, 8, 8. [Google Scholar]
- Pawlowski, A.; Jansson, M.; Sköld, M.; Rottenberg, M.E.; Källenius, G. Tuberculosis and HIV Co-Infection. PLoS Pathog 2012, 8, e1002464. [Google Scholar] [CrossRef]
- Shyam, M.; Shilkar, D.; Rakshit, G.; Jayaprakash, V. Approaches for Targeting the Mycobactin Biosynthesis Pathway for Novel Anti-Tubercular Drug Discovery: Where We Stand. Expert Opin Drug Discov 2022, 17, 699–715. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiology and molecular biology reviews 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Quadri, L.E.N. Iron Uptake in Mycobacteria. The mycobacterial cell envelope 2008, 167–184. [Google Scholar]
- Hammer, N.D.; Skaar, E.P. Molecular Mechanisms of Staphylococcus Aureus Iron Acquisition. Annu Rev Microbiol 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.; Ewing, D.F.; Ratledge, C. Isolation, Identification, and Structural Analysis of the Mycobactins of Mycobacterium Avium, Mycobacterium Intracellulare, Mycobacterium Scrofulaceum, and Mycobacterium Paratuberculosis. J Bacteriol 1985, 164, 896–903. [Google Scholar] [CrossRef]
- Hall, R.M.; Ratledge, C. A Simple Method for the Production of Mycobactin, the Lipid-Soluble Siderophore, from Mycobacteria. FEMS Microbiol Lett 1982, 15, 133–136. [Google Scholar] [CrossRef]
- De Voss, J.J.; Rutter, K.; Schroeder, B.G.; Su, H.; Zhu, Y.; Barry III, C.E. The Salicylate-Derived Mycobactin Siderophores of Mycobacterium Tuberculosis Are Essential for Growth in Macrophages. Proceedings of the National Academy of Sciences 2000, 97, 1252–1257. [Google Scholar] [CrossRef]
- Barry, S.M.; Challis, G.L. Recent Advances in Siderophore Biosynthesis. Curr Opin Chem Biol 2009, 13, 205–215. [Google Scholar] [CrossRef]
- Snow, G. Mycobactins: Iron-Chelating Growth Factors from Mycobacteria. Bacteriol Rev 1970, 34, 99–125. [Google Scholar] [CrossRef]
- Sritharan, M. Iron Homeostasis in Mycobacterium Tuberculosis: Mechanistic Insights into Siderophore-Mediated Iron Uptake. J Bacteriol 2016, 198, 2399–2409. [Google Scholar] [CrossRef]
- Wells, R.M.; Jones, C.M.; Xi, Z.; Speer, A.; Danilchanka, O.; Doornbos, K.S.; Sun, P.; Wu, F.; Tian, C.; Niederweis, M. Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium Tuberculosis. PLoS Pathog 2013, 9, e1003120. [Google Scholar] [CrossRef]
- Choudhury, M.; Koduru, T.N.; Kumar, N.; Salimi, S.; Desai, K.; Prabhu, N.P.; Sritharan, M. Iron Uptake and Transport by the Carboxymycobactin-Mycobactin Siderophore Machinery of Mycobacterium Tuberculosis Is Dependent on the Iron-Regulated Protein HupB. BioMetals 2021, 34, 511–528. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Smith, I. Identification of an ABC Transporter Required for Iron Acquisition and Virulence in Mycobacterium Tuberculosis. J Bacteriol 2006, 188, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Ryndak, M.B.; Wang, S.; Smith, I.; Rodriguez, G.M. The Mycobacterium Tuberculosis High-Affinity Iron Importer, IrtA, Contains an FAD-Binding Domain. J Bacteriol 2010, 192, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Gupta, R.K.; Khare, G.; Salunke, D.M.; Tyagi, A.K. Crystal Structure of Bfr A from Mycobacterium Tuberculosis: Incorporation of Selenomethionine Results in Cleavage and Demetallation of Haem. PLoS ONE 2009, 4, e8028. [Google Scholar] [CrossRef]
- Yao, H.; Wang, Y.; Lovell, S.; Kumar, R.; Ruvinsky, A.M.; Battaile, K.P.; Vakser, I.A.; Rivera, M. The Structure of the BfrB–Bfd Complex Reveals Protein–Protein Interactions Enabling Iron Release from Bacterioferritin. J Am Chem Soc 2012, 134, 13470–13481. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, M.S.; Unnikrishnan, M.; McConnell, M.J.; Borowsky, M.; Cheng, T.-Y.; Siddiqi, N.; Fortune, S.M.; Moody, D.B.; Rubin, E.J. Mycobacterial Esx-3 Is Required for Mycobactin-Mediated Iron Acquisition. Proceedings of the National Academy of Sciences 2009, 106, 18792–18797. [Google Scholar] [CrossRef] [PubMed]
- Serafini, A.; Pisu, D.; Palù, G.; Rodriguez, G.M.; Manganelli, R. The ESX-3 Secretion System Is Necessary for Iron and Zinc Homeostasis in Mycobacterium Tuberculosis. PLoS ONE 2013, 8, e78351. [Google Scholar] [CrossRef]
- Jones, C.M.; Wells, R.M.; Madduri, A.V.R.; Renfrow, M.B.; Ratledge, C.; Moody, D.B.; Niederweis, M. Self-Poisoning of Mycobacterium Tuberculosis by Interrupting Siderophore Recycling. Proceedings of the National Academy of Sciences 2014, 111, 1945–1950. [Google Scholar] [CrossRef]
- Shyam, M.; Verma, H.; Bhattacharje, G.; Mukherjee, P.; Singh, S.; Kamilya, S.; Jalani, P.; Das, S.; Dasgupta, A.; Mondal, A. Mycobactin Analogues with Excellent Pharmacokinetic Profile Demonstrate Potent Antitubercular Specific Activity and Exceptional Efflux Pump Inhibition. J Med Chem 2022, 65, 234–256. [Google Scholar] [CrossRef] [PubMed]
- Stirrett, K.L.; Ferreras, J.A.; Jayaprakash, V.; Sinha, B.N.; Ren, T.; Quadri, L.E.N. Small Molecules with Structural Similarities to Siderophores as Novel Antimicrobials against Mycobacterium Tuberculosis and Yersinia Pestis. Bioorg Med Chem Lett 2008, 18, 2662–2668. [Google Scholar] [CrossRef]
- Ferreras, J.A.; Gupta, A.; Amin, N.D.; Basu, A.; Sinha, B.N.; Worgall, S.; Jayaprakash, V.; Quadri, L.E.N. Chemical Scaffolds with Structural Similarities to Siderophores of Nonribosomal Peptide-Polyketide Origin as Novel Antimicrobials against Mycobacterium Tuberculosis and Yersinia Pestis. Bioorg Med Chem Lett 2011, 21, 6533–6537. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15–Ligand Discovery for Everyone. J Chem Inf Model 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Irwin, J.J.; Tang, K.G.; Young, J.; Dandarchuluun, C.; Wong, B.R.; Khurelbaatar, M.; Moroz, Y.S.; Mayfield, J.; Sayle, R.A. ZINC20—A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J Chem Inf Model 2020, 60, 6065–6073. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Huey, R.; Hart, W.E.; Halliday, S.; Belew, R.; Olson, A.J. AutoDock. Automated docking of flexible ligands to receptor-User Guide, 2001. [Google Scholar]
- Rizvi, S.M.D.; Shakil, S.; Haneef, M. A Simple Click by Click Protocol to Perform Docking: Autodock 4.2 Made Easy for Non-Bioinformaticians. EXCLI J 2013, 12, 830–857. [Google Scholar]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J Comput Chem 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr 2002, 40, 82–92. [Google Scholar]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery 2011.
- Irwin, J.J.; Shoichet, B.K. ZINC− a Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model 2005, 45, 177–182. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J Cheminform 2011, 3, 1–14. [Google Scholar] [CrossRef]
- May, J.J.; Kessler, N.; Marahiel, M.A.; Stubbs, M.T. Crystal Structure of DhbE, an Archetype for Aryl Acid Activating Domains of Modular Nonribosomal Peptide Synthetases. Proceedings of the National Academy of Sciences 2002, 99, 12120–12125. [Google Scholar] [CrossRef]
- Vergnolle, O.; Xu, H.; Tufariello, J.M.; Favrot, L.; Malek, A.A.; Jacobs, W.R.; Blanchard, J.S. Post-Translational Acetylation of MbtA Modulates Mycobacterial Siderophore Biosynthesis. Journal of Biological Chemistry 2016, 291, 22315–22326. [Google Scholar] [CrossRef] [PubMed]
- Schieffer, G.; Peng, I. Accelerating Drug Discovery in AutoDock-GPU with Tensor Cores. In Proceedings of the European Conference on Parallel Processing; Springer; 2023; pp. 608–622. [Google Scholar]
- Kalibaeva, G.; Ferrario, M.; Ciccotti, G. Constant Pressure-Constant Temperature Molecular Dynamics: A Correct Constrained NPT Ensemble Using the Molecular Virial. Mol Phys 2003, 101, 765–778. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. Protein dynamics: Methods and protocols 2014, 193–226. [Google Scholar]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential Dynamics of Proteins. Proteins: Structure, Function, and Bioinformatics 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Deeks, E.D. Cobicistat: A Review of Its Use as a Pharmacokinetic Enhancer of Atazanavir and Darunavir in Patients with HIV-1 Infection. Drugs 2014, 74, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.; Faulds, D. Saquinavir: A Review of Its Pharmacology and Clinical Potential in the Management of HIV Infection. Drugs 1996, 52, 93–112. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.L.; Stewart, A.K. Carfilzomib: A Novel Second-Generation Proteasome Inhibitor. Future oncology 2011, 7, 607–612. [Google Scholar] [CrossRef]
- Husain, A.; Azim, M.S.; Mitra, M.; Bhasin, P.S. A Review on Candesartan: Pharmacological and Pharmaceutical Profile. J Appl Pharm Sci 2011, 12–17. [Google Scholar]
- Mangum, E.M.; Graham, K.K. Lopinavir-ritonavir: A New Protease Inhibitor. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 2001, 21, 1352–1363. [Google Scholar] [CrossRef]
- Plosker, G.L.; Noble, S. Indinavir: A Review of Its Use in the Management of HIV Infection. Drugs 1999, 58, 1165–1203. [Google Scholar] [CrossRef]
- Chung, K.F.; Barnes, P.J. Zafirlukast (Accolate). Drugs Today (Barc) 1998, 34, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.R.; Mishra, P.; Abraham, J. Neratinib, a Novel HER2-Targeted Tyrosine Kinase Inhibitor. Clin Breast Cancer 2016, 16, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Lim-Wilby, M. Molecular Docking. Molecular modeling of proteins 2008, 365–382. [Google Scholar]
- Labello, N.P.; Bennett, E.M.; Ferguson, D.M.; Aldrich, C.C. Quantitative Three Dimensional Structure Linear Interaction Energy Model of 5′-O-[N-(Salicyl) Sulfamoyl] Adenosine and the Aryl Acid Adenylating Enzyme MbtA. J Med Chem 2008, 51, 7154–7160. [Google Scholar] [CrossRef] [PubMed]

| AutoDock 4.2.6 | |||||||
|---|---|---|---|---|---|---|---|
| Protein |
Center grid box dimensions (Å) |
Spacing (Å) | Coordinates for the center of the grid box | ||||
| X-axis | Y-axis | Z-axis | 0.375 | X-axis | Y-axis | Z-axis | |
| MbtA | 50 | 50 | 50 | -2.439 | 19.515 | 32.895 | |
| S. No | Code | Structure | Compounds Details | MOA and Therapeutic application |
|---|---|---|---|---|
| 1 | a_617 |  |
ZINC ID: ZINC000085537014 Cobicistat Formula: C40H53N7O5S2 Molar mass: 776.03 g·mol−1 |
Cobicistat (Tybost™) functions as a mechanism-based inhibitor of cytochrome P450 (CYP) 3A enzymes. It is approved in the EU as a pharmacokinetic enhancer, specifically a booster, of HIV-1 protease inhibitors (PIs) such as atazanavir and darunavir in adult patients[49] |
| 2 | a_391 | 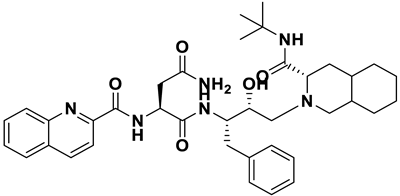 |
ZINC ID: ZINC000003914596 Saquinavir Formula: C38H50N6O5 Molar mass: 670.855 g·mol−1 |
Saquinavir was the first protease inhibitor developed for HIV therapy. It acts by cleaving viral polypeptide chains into functional proteins during the late stages of HIV replication[50] |
| 3 | a_821 |  |
ZINC ID: ZINC000049841054 Carfilzomib Formula: C38H50N6O5 Molar mass: 670.855 g·mol−1 |
Carfilzomib (formerly PR-171) is a novel epoxyketone-based irreversible proteasome inhibitor. It specifically targets the chymotrypsin-like activity of the proteasome, preventing its normal function and causing a buildup of proteins within the cancer cells[51]. |
| 4 | a_827 |  |
ZINC ID: ZINC000004074875 Candesartan Formula: C24H20N6O3 Molar mass: 440.463 g·mol−1 |
Candesartan blocks the vasoconstrictor and aldosterone-secreting effects of angiotensin II by selectively blocking the binding of angiotensin II to the AT1 receptor in many tissues, such as vascular smooth muscle and the adrenal gland[52]. |
| 5 | a_85 |  |
ZINC ID: ZINC000003944422 Ritonavir Formula: C37H48N6O5S2 Molar mass: 720.95 g·mol−1 |
Ritonavir is a protease inhibitor and is used as booster with other protease inhibitors in the treatment of HIV infection. It prevents the proper maturation of the virus[53]. |
| 6 | a_472 | 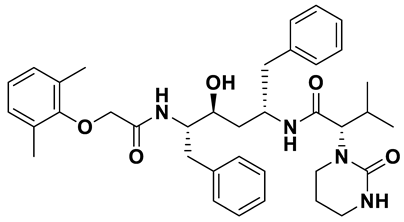 |
ZINC ID: ZINC000003951740 Lopinavir Formula: C37H48N4O5 Molar mass: 628.814 g·mol−1 |
Lopinavir is an antiretroviral medication primarily used in the treatment of HIV infection. It acts as a protease inhibitor in the final stages of the viral replication process thereby preventing the proper maturation of the virus leading to immature and non-functional viral particles[53]. |
| 7 | a_1276 | 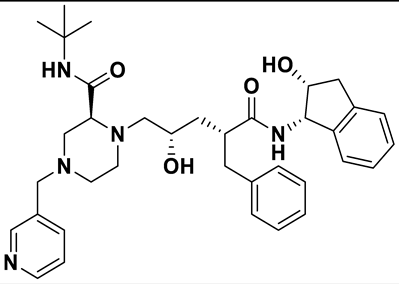 |
ZINC ID: ZINC000022448696 Indinavir Formula: C36H47N5O4 Molar mass: 613.803 g·mol−1 |
Indinavir is a protease inhibitor used as a component of highly active antiretroviral therapy to treat HIV/AIDS. It interferes with viral maturation, and contributes to the control of viral replication. It is also used as booster with other protease inhibitors[54]. |
| 8 | a_1338 | 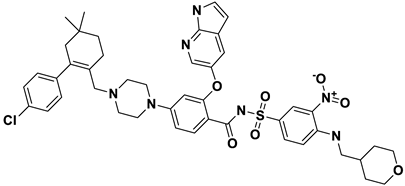 |
ZINC ID: ZINC000150338755 Venetoclax Formula: C45H50ClN7O7S Molar mass: 867.320 g·mol−1 |
Venetoclax is B-cell lymphoma-2 (BCL-2) inhibitors. It binds to the BCL-2 protein, blocking its anti-apoptotic function |
| 9 | a_797 | 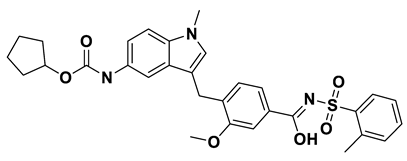 |
ZINC ID: ZINC000000896717 Accolate Formula: C31H33N3O6S Molar mass: 575.210 g·mol−1 |
Zafirlukast (Accolate) is a leukotriene receptor antagonist. It inhibits the activity of leukotrienes by binding to their receptors found on various cells in the airways causing bronchodilation[55]. |
| 10 | a_1388 | 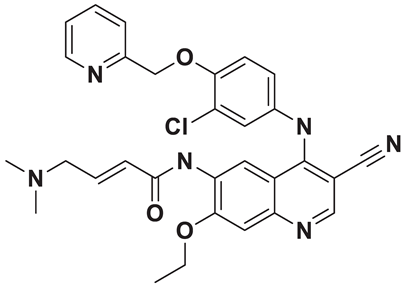 |
ZINC ID: ZINC000003916214 Neratinib Formula: C30H29ClN6O3 Molar mass: 556.200 g·mol−1 |
Neratinib is a tyrosine kinase inhibitor used in the treatment of certain types of breast cancers (targeted therapy). It works by irreversibly inhibiting multiple receptor tyrosine kinases, including HER2, epidermal growth factor receptor (EGFR), and HER4[56]. |
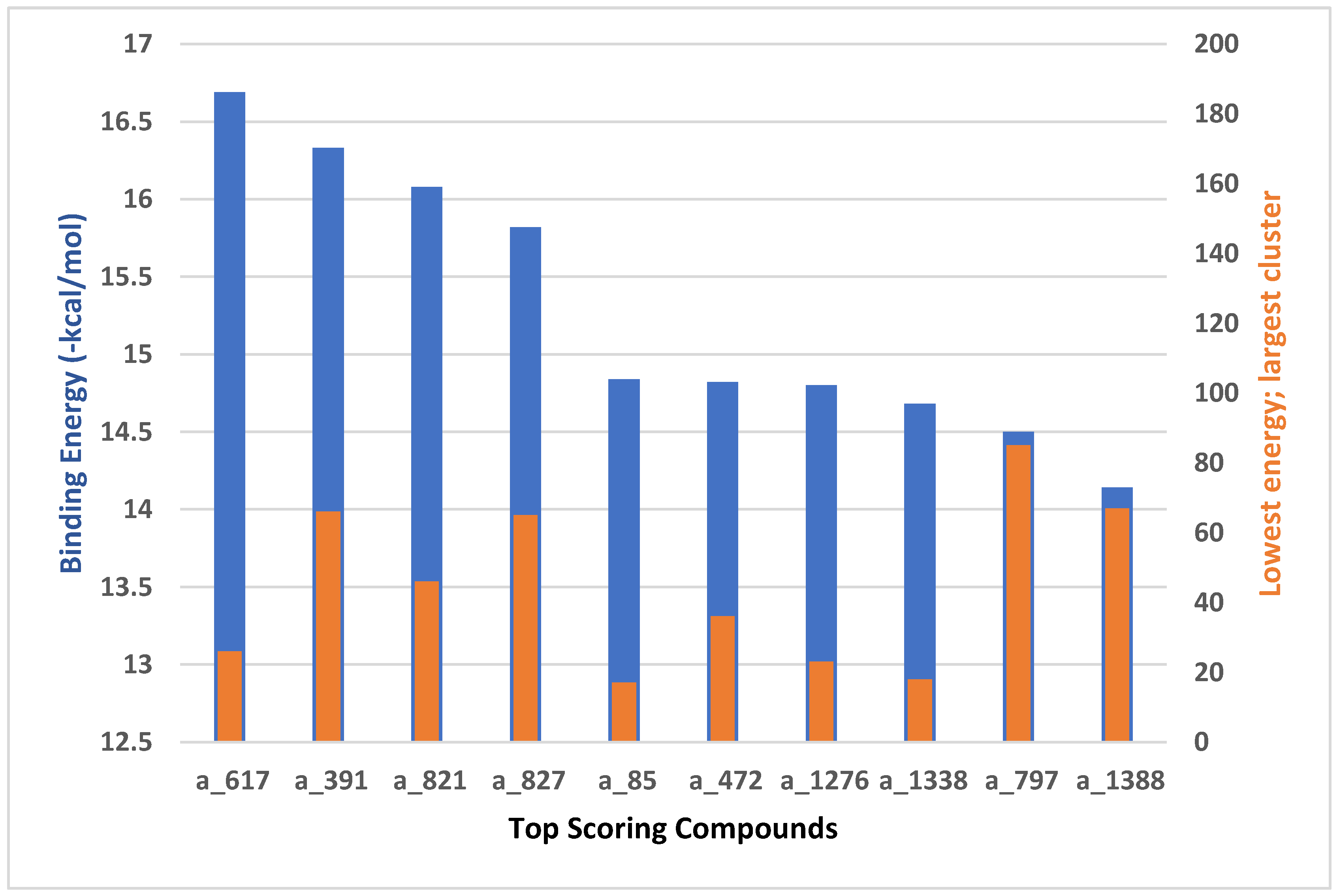
| S. No | Ligand | Runs | Binding Energy (kcal/mol) | ESTAT | HB | VDW | DSOLV |
|---|---|---|---|---|---|---|---|
| 1 | a_617 | 200 | -16.69 | -0.9438 | -2.5163 | -23.9398 | 5.5597 |
| 2 | a_391 | 200 | -16.33 | -0.6982 | -2.18 | -21.8025 | 4.9488 |
| 3 | a_821 | 200 | -16.08 | -1.1546 | -1.562 | -23.9542 | 5.2432 |
| 4 | a_827 | 200 | -15.82 | -0.8058 | -1.9775 | -21.7725 | 4.9785 |
| 5 | a_85 | 200 | -14.84 | -0.3628 | -2.1603 | -20.6416 | 4.0052 |
| 6 | a_472 | 200 | -14.82 | -0.8465 | -2.6141 | -20.8497 | 5.0782 |
| 7 | a_1276 | 200 | -14.80 | -0.9278 | -2.6608 | -20.6792 | 5.1776 |
| 8 | a_1338 | 200 | -14.68 | -1.3963 | -1.6507 | -27.4664 | 6.791 |
| 9 | a_797 | 200 | -14.50 | -0.0758 | -1.9036 | -22.3757 | 4.7207 |
| 10 | a_1388 | 200 | -14.14 | -0.0544 | -1.7261 | -21.6482 | 4.6123 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





