Submitted:
15 September 2023
Posted:
19 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
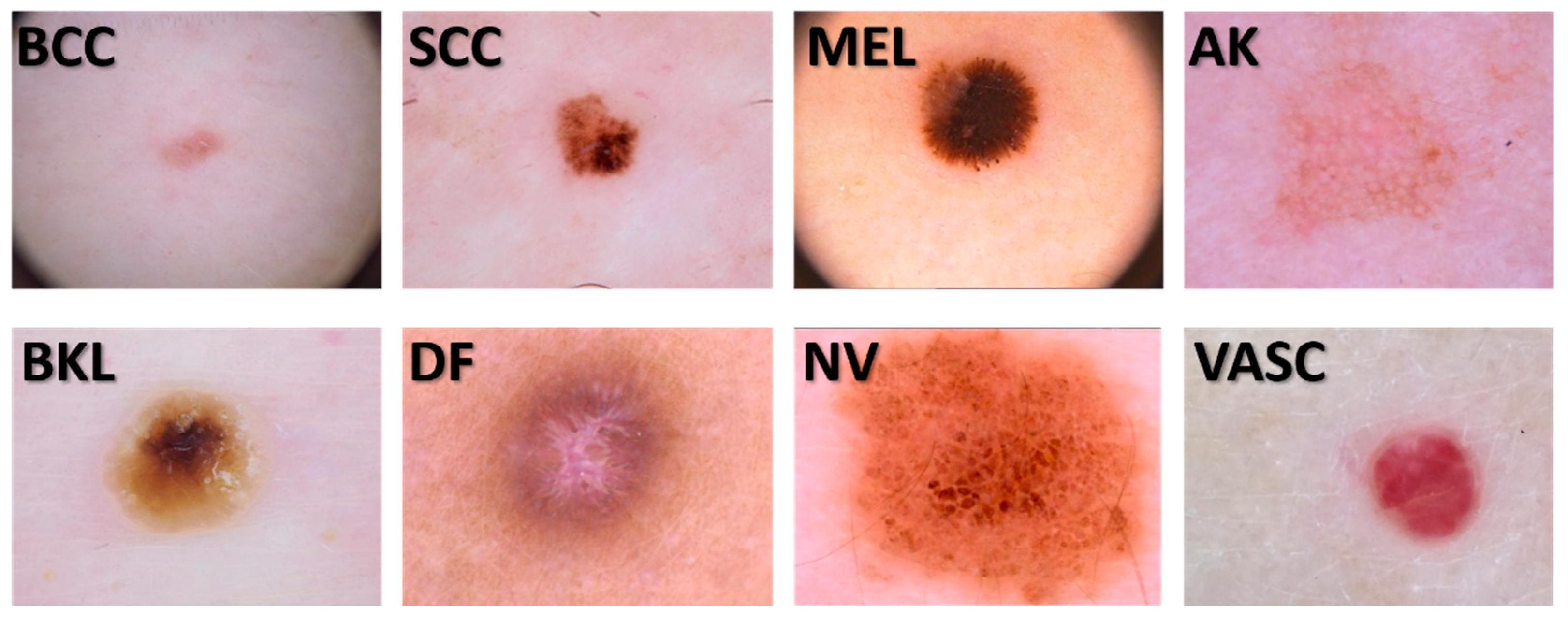
2.1. Image dataset
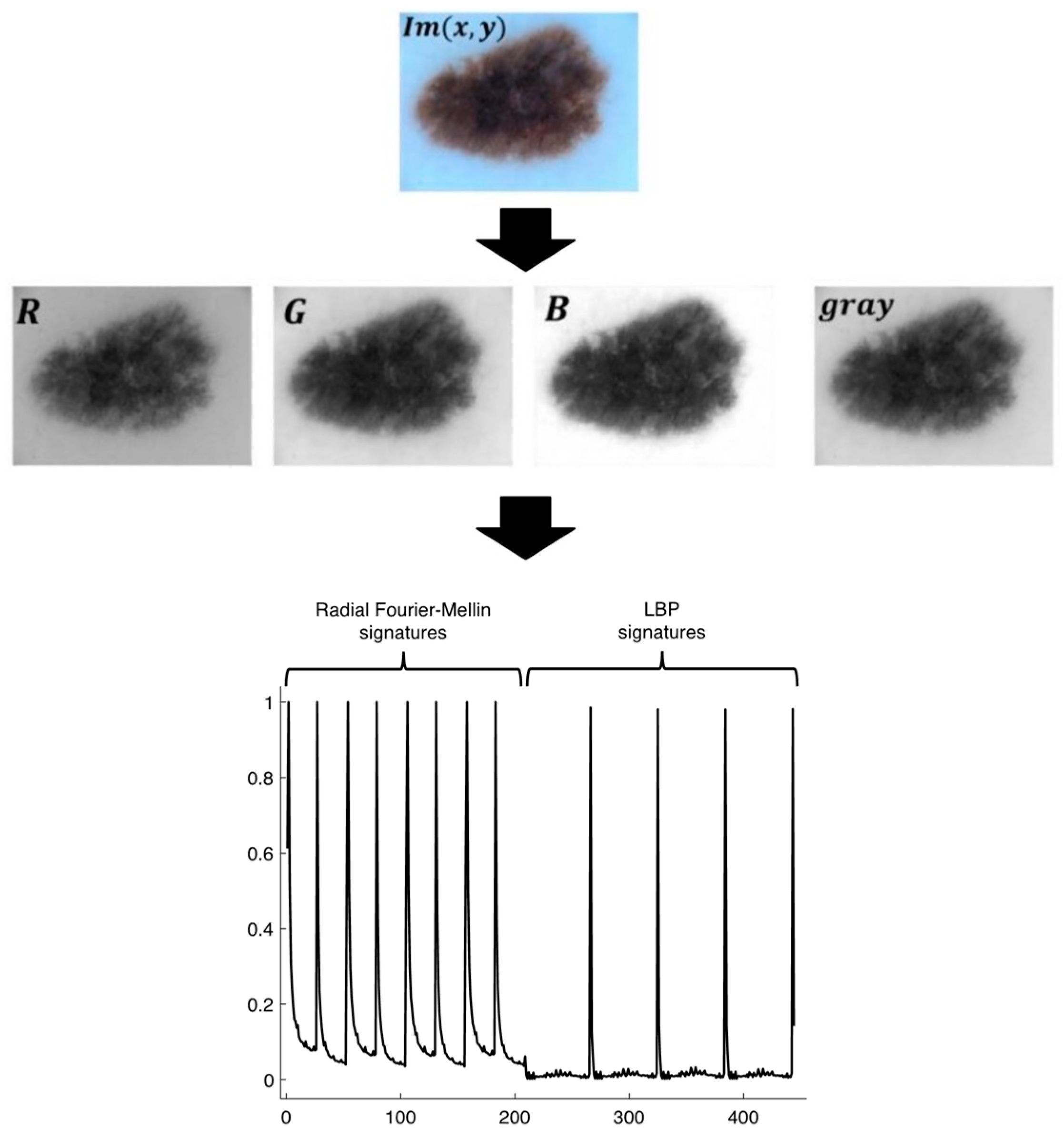
2.2. The signatures
2.3. Radial Fourier-Mellin signatures through Hilbert transform
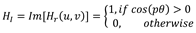
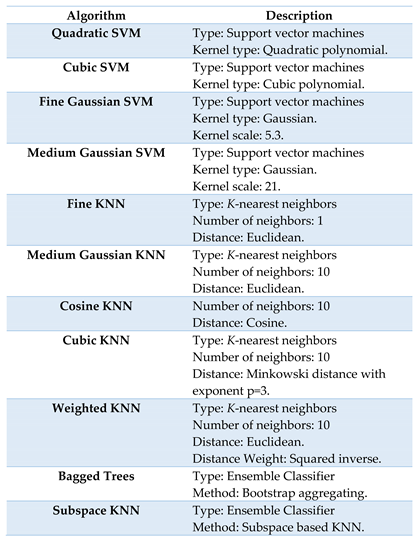
2.4. Signature Classification
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lomas, A.; Leonardi-Bee, J.; Bath-Hextall, F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012, 166, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R. Skin cancer: an overview of epidemiology and risk factors. Semin Oncol Nurs. 2013, 29, 160–169. [Google Scholar] [CrossRef]
- Cameron, M.C.; Lee, E.; Hibler, B.P.; Barker, C.A.; Mori, S.; Cordova, M.; Nehal, K.S.; Rossi, A.M. Basal cell carcinoma: epidemiology; pathophysiology; clinical and histological subtypes; and disease associations. J Am Acad Dermatol. 2019, 80, 303–317. [Google Scholar] [CrossRef]
- Zhang, W.; Zeng, W.; Jiang, A.; He, Z.; Shen, X.; Dong, X.; Feng, J.; Lu, H. Global, regional and national incidence, mortality and disability-adjusted life-years of skin cancers and trend analysis from 1990 to 2019: An analysis of the Global Burden of Disease Study 2019. Cancer Med. 2021, 10, 4905–4922. [Google Scholar] [CrossRef] [PubMed]
- Born, L.J. , Khachemoune, A. Basal cell carcinosarcoma: a systematic review and reappraisal of its challenges and the role of Mohs surgery. Arch Dermatol Res, 2023, 315, 2195–2205. [Google Scholar] [CrossRef]
- Naik, P.P.; Desai, M.B. Basal Cell Carcinoma: A Narrative Review on Contemporary Diagnosis and Management. Oncol Ther, 2022, 10, 317–335. [Google Scholar] [CrossRef]
- Reinehr, C.P.H.; Bakos, R.M. Actinic keratoses: review of clinical, dermoscopic, and therapeutic aspects. An Bras Dermatol. 2019, 94, 637–657. [Google Scholar] [CrossRef]
- Del Regno, L.; Catapano, S.; Di Stefani, A.; Cappilli, S.; Peris, K. A Review of Existing Therapies for Actinic Keratosis: Current Status and Future Directions. Am J Clin Dermatol, 2022, 23, 339–352. [Google Scholar] [CrossRef]
- Casari, A.; Chester, J.; Pellacani, G. Actinic Keratosis and Non-Invasive Diagnostic Techniques: An Update. Biomedicines, 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Opoko, U.; Sabr, A.; Raiteb, M.; Maadane, A.; Slimani, F. Seborrheic keratosis of the cheek simulating squamous cell carcinoma. Int J Surg Case Rep. 2021, 84. [Google Scholar] [CrossRef]
- Moscarella, E.; Brancaccio, G.; Briatico, G.; Ronchi, A.; Piana, S. Argenziano G. Differential Diagnosis and Management on Seborrheic Keratosis in Elderly Patients. Clin Cosmet Investig Dermatol. 2021, 14, 395–406. [Google Scholar] [CrossRef]
- Jiahua, Xing.; Yi, Chen.; Liwu, Zheng.; Yan, Shao.; Yichi, Xu.; Lingli, Guo. Innovative combined therapy for multiple keloidal dermatofibromas of the chest wall: A novel case report. CJPRS, 2022, 4, 182–186. [CrossRef]
- Sofia, Endzhievskaya.; Chao-Kai, Hsu.; Hsing-San, Yang.; Hsin-Yu, Huang.; Yu-Chen, Lin.; Yi-Kai, Hong.; John, Y.W. Lee.; Alexandros, Onoufriadis.; Takuya, Takeichi,.; Julia, Yu-Yun, Lee.; Tanya, J. Shaw.; John, A.McGrath.; Maddy, Parsons. Loss of RhoE Function in Dermatofibroma Promotes Disorganized Dermal Fibroblast Extracellular Matrix and Increased Integrin Activation. JID 2023, 143, 1487–1497. [CrossRef]
- Park, S.; Yun, S.J. Acral Melanocytic Neoplasms: A Comprehensive Review of Acral Nevus and Acral Melanoma in Asian Perspective. Dermatopathology 2022, 9, 292–303. [Google Scholar] [CrossRef]
- Frischhut, N.; Zelger, B.; Andre, F.; Zelger, B.G. The spectrum of melanocytic nevi and their clinical implications. J Dtsch Dermatol Ges. 2022, 20, 483–504. [Google Scholar] [CrossRef]
- Hu, K.; Li, Y.; Ke, Z.; Yang, H.; Lu, C.; Li, Y.; Guo, Y.; Wang, W. History, progress and future challenges of artificial blood vessels: a narrative review. Biomater Transl. 2022, 28, 81–98. [Google Scholar] [CrossRef]
- Liu, C.; Dai, J.; Wang, X.; Hu, X. The Influence of Textile Structure Characteristics on the Performance of Artificial Blood Vessels. Polymers. 2023, 15, 1–24. [Google Scholar] [CrossRef]
- Folland, G.B. Fourier Analysis and Its Applications; American Mathematical Society: Providence, RI, USA, 2000; pp. 314–318. [Google Scholar]
- Al-masni, M.A.; Al-antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Meth. Prog. Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef]
- Afza, F.; Sharif, M.; Khan, M.A.; Tariq, U.; Yong, H.-S.; Cha, J. Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine. Sensors. 2022, 22, 799. [Google Scholar] [CrossRef] [PubMed]
- Shetty, B.; Fernandes, R.; Rodrigues, A. P.; Chengoden, R.; Bhattacharya, S.; Lakshmanna, K. Skin lesion classification of dermoscopic images using machine learning and convolutional neural network. Scientific reports. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Camacho-Gutiérrez, J. A.; Solorza-Calderón, S.; Álvarez-Borrego, J. Multi-class skin lesion classification using prism- and segmentation-based fractal signatures. Expert Syst. Appl. 2022, 197. [Google Scholar] [CrossRef]
- Moldovanu, S.; Damian Michis, F.A.; Biswas, K.C.; Culea-Florescu, A.; Moraru, L. Skin Lesion Classification Based on Surface Fractal Dimensions and Statistical Color Cluster Features Using an Ensemble of Machine Learning Techniques. Cancers 2021, 13. [Google Scholar] [CrossRef]
- Afza, F.; Khan, M.A.; Sharif, M.; Saba, T.; Rehman, A.; Javed, M.Y. Skin Lesion Classification: An Optimized Framework of Optimal Color Features Selection. 2020 2nd International Conference on Computer and Information Sciences (ICCIS), Sakaka, Saudi Arabia, 2020. [CrossRef]
- Ghalejoogh, G.S.; Kordy, H.M.; Ebrahimi, F. A hierarchical structure based on Stacking approach for skin lesion classification. Expert Syst. Appl. 2020, 145. [Google Scholar] [CrossRef]
- Fisher, R.; Rees, J.; Bertrand, A. Classification of Ten Skin Lesion Classes: Hierarchical KNN versus Deep Net. in Medical Image Understanding and Analysis: 23rd Conference, MIUA 2019, Liverpool, UK, July 24-26, 2019, Proceedings. Communications in Computer and Information Science (CCIS), Springer 2020. 24 July. [CrossRef]
- Lynn, N. C.; Kyu, Z. M. Segmentation and Classification of Skin Cancer Melanoma from Skin Lesion Images. 18th International Conference on Parallel and Distributed Computing, Applications and Technologies (PDCAT), 2017. [CrossRef]
- DermWeb. Available online: http://www.dermweb.com/dull_razor/ (accessed on 7 April 1997).
- Al-abayechi, A.A.; Jalab, H.A.; Ibrahim, R.W. A classification of skin lesion using fractional poisson for texture feature extraction. Proceedings of the Second International Conference on Internet of things, Data and Cloud Computing 2017. [CrossRef]
- Ozkan, I.A.; Koklu, M. Skin Lesion Classification using Machine Learning Algorithms. IJISAE. 2017, 5, 285–289. [Google Scholar] [CrossRef]
- Chakravorty, R.; Liang, S.; Abedini, M.; Garnavi, R. Dermatologist-like feature extraction from skin lesion for improved asymmetry classification in PH2 database. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016. [Google Scholar] [CrossRef]
- Leo, C.D.; Bevilacqua, V.; Ballerini, L.; Fisher, R.B.; Aldridge, B.; Rees, J. Hierarchical Classification of Ten Skin Lesion Classes. In Proceedings of the SICSA Dundee Medical Image Analysis Workshop, Dundee, UK, 27 March 2015. [Google Scholar]
- Patil, P. P.; Patil, S. A.; Udupi, V. Detection and classification of skin lesion in dermoscopy images. Int. J. Appl. Eng. Res. 2014, 9, 27719–27731. [Google Scholar]
- Cavalcanti, P. G.; Scharcanski, J. Macroscopic pigmented skin lesion segmentation and its influence on lesion classification and diagnosis. In Color Medical Image Analysis. Lecture Notes in Computational Vision and Biomechanics; Celebi, M., Schaefer, G., Eds.; Dordrecht: Springer Netherlands, 2013; Volume 6, pp. 15–39. [Google Scholar] [CrossRef]
- Ballerini, L.; Fisher, R. B.; Aldridge, B.; Rees, J. Non-melanoma skin lesion classification using colour image data in a hierarchical k-nn classifier. In Proceedings of the 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 05 May 2012. [Google Scholar] [CrossRef]
- Ramlakhan, K.; Shang, Y. A mobile automated skin lesion classification system. In Proceedings of the 2011 IEEE 23rd International Conference on Tools with Artificial Intelligence, Boca Raton, FL, USA, 7–9 November 2011. [Google Scholar] [CrossRef]
- Mohanty, N.; Pradhan, M.; Reddy, A. V. N.; Kumar, S.; Alkhayyat, A. Integrated Design of Optimized Weighted Deep Feature Fusion Strategies for Skin Lesion Image Classification. Cancers. 2022, 14, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Benyahia, S.; Meftah, B.; Lézoray, O. Multi-features extraction based on deep learning for skin lesion classification. Tissue Cell. 2022, 74, 101701. [Google Scholar] [CrossRef] [PubMed]
- Melbin, K.; Raj, Y.J.V. Integration of modified ABCD features and support vector machine for skin lesion types classification. Multimed. Tools. Appl. 2021, 80, 8909–8929. [Google Scholar] [CrossRef]
- Guha, S.R.; Rafizul Haque, S.M. Performance Comparison of Machine Learning-Based Classification of Skin Diseases from Skin Lesion Images. In International Conference on Communication, Computing and Electronics Systems. Lecture Notes in Electrical Engineering; Bindhu, V., Chen, J., Tavares, J., Eds.; Springer: Singapore, 2020; Volume 637, pp. 15–25. [Google Scholar] [CrossRef]
- Chatterjee, S.; Dey, D.; Munshi, S. Integration of morphological preprocessing and fractal based feature extraction with recursive feature elimination for skin lesion types classification. Comput. Methods. Program.s Biomed. 2019, 178, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Javed, R.; Saba, T.; Shafry, M.; Rahim, M. An Intelligent Saliency Segmentation Technique and Classification of Low Contrast Skin Lesion Dermoscopic Images Based on Histogram Decision. In Proceedings of the 2019 12th International Conference on Developments in eSystems Engineering (DeSE), Kazan, Russia, 7–10 October 2019. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Saba, T.; Javed, K.; Lali, I.U.; Tanik, U.J. Rehman, A. Construction of saliency map and hybrid set of features for efficient segmentation and classification of skin lesion. Microsc. Res. Tech. 2019, 82, 741–763. [Google Scholar] [CrossRef]
- Filali, Y.; Abdelouahed, S.; Aarab, A. An Improved Segmentation Approach for Skin Lesion Classification. Stat. Optim. Inf. Comput. 2019, 7, 456–467. [Google Scholar] [CrossRef]
- Akram, T.; Khan, M.A.; Sharif, M.; Yasmin, M. Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J. Ambient. Intell. Human. Comput. 2018, 1–20. [Google Scholar] [CrossRef]
- Nasir, M.; Attique Khan, M.; Sharif, M.; Lali, I. U.; Saba, T.; Iqbal, T. An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microsc. Res. Tech. 2018, 81, 528–543. [Google Scholar] [CrossRef]
- Wahba, M.A.; Ashour, A.S.; Guo, Y.; Napoleon, S.A.; Abd-Elnaby, M.M. A novel cumulative level difference mean based GLDM and modified ABCD features ranked using eigenvector centrality approach for four skin lesion types classification. Comput. Methods. Programs. Biomed. 2018, 165, 163–174. [Google Scholar] [CrossRef]
- Wahba, M. A.; Ashour, A. S.; Napoleon, S. A.; Abd Elnaby, M. M.; Guo, Y. Combined empirical mode decomposition and texture features for skin lesion classification using quadratic support vector machine. Health. Inf. Sci. Syst. 2017, 5, 1–13. [Google Scholar] [CrossRef]
- Satheesha, T. Y.; Satyanarayana, D.; Prasad, M. N. G.; Dhruve, K. D. Melanoma Is Skin Deep: A 3D Reconstruction Technique for Computerized Dermoscopic Skin Lesion Classification. IEEE J. Transl. Eng. Health. Med. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Danpakdee, N.; Songpan, W. Classification Model for Skin Lesion Image. In Information Science and Applications 2017. ICISA 2017. Lecture Notes in Electrical Engineering, Kim, K., Joukov, N., Eds.; Springer: Singapore, 2017; Volume 424, pp. 553–561. [Google Scholar] [CrossRef]
- Filali, Y.; Ennouni, A.; Sabri, M. A.; Aarab, A. Multiscale approach for skin lesion analysis and classification. In Proceedings of the 2017 International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Fez, Morocco, 22–24 May 2017. [Google Scholar] [CrossRef]
- Farooq, M.A.; Azhar, M.A.; Raza, R.H. Automatic Lesion Detection System (ALDS) for Skin Cancer Classification Using SVM and Neural Classifiers. In Proceedings of the 2016 IEEE 16th International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 31 October 2016–02 November 2016. [Google Scholar] [CrossRef]
- Sirakov, N.M.; Ou, YL.; Mete, M. Skin lesion feature vectors classification in models of a Riemannian manifold. Ann. Math. Artif. Intell. 2015, 75, 217–229. [Google Scholar] [CrossRef]
- Mete, M.; Ou, YL.; Sirakov, N.M. Skin Lesion Feature Vector Space with a Metric to Model Geometric Structures of Malignancy for Classification. In Combinatorial Image Analaysis. IWCIA 2012. Lecture Notes in Computer Science, Barneva, R.P., Brimkov, V.E., Aggarwal, J.K., Eds.; Springer, Berlin, Heidelberg, Germany, 2012; Volume 7655, pp. 285–297. [CrossRef]
- Amelard, R.; Wong, A.; Clausi, D. A. Extracting morphological high-level intuitive features (HLIF) for enhancing skin lesion classification. In 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August 2012 - 01 September 2012. 28 August. [CrossRef]
- Molina-Molina, E.O.; Solorza-Calderón, S.; Álvarez-Borrego, J. Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features. Appl. Sci. 2020, 10, 5954. [Google Scholar] [CrossRef]
- Surówka, G; Ogorzalek, M. On optimal wavelet bases for classification of skin lesion images through ensemble learning. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 06-11 July 2014. [CrossRef]
- López-Ávila, L. F.; Álvarez-Borrego, J.; Solorza-Calderón, S. Fractional Fourier-Radial Transform for Digital Image Recognition. J. Sign. Process. Syst. 2021, pp. 49–66 (2021). [CrossRef]
- ISICCHALLENGE. Available online: https://challenge.isic-archive.com/data/#2019.
- Casasent, D. , Psaltis, D. Scale invariant optical correlation using Mellin transforms. Opt. Commun. 1976, 17, 59–63. [Google Scholar] [CrossRef]
- Derrode, S. , Ghorbel, F. Robust and efficient Fourier– Mellin transform approximations for gray-level image reconstruction and complete invariant description. Comput. Vis. Image. Underst. 2001, 83, 57–78. [Google Scholar] [CrossRef]
- Alcaraz-Ubach, D.F. Reconocimiento de patrones en imágenes digitales usando máscaras de Hilbert Binarias de anillos concéntricos. Bachelor thesis, Science Faculty, UABC, Ensenada, Baja California, México, 2015.
- Davis, J. A., McNamara, D. E., Cottrell, D. M., Campos, J. Image processing with the radial Hilbert transform: Theory and experiments. Opt. Lett. 2000, 25, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.C., Ding, J.J. The generalized radial Hilbert transform and its applications to 2D edge detection (any direction or specified directions). In Proceedings of the 2003 IEEE International Conference on Acoustics, Speech, and Signal, Hong Kong China, 06-10 April 2003. Speech, and Signal, Hong Kong China, 6–10 April 2003. [CrossRef]
- King, F. W. Hilbert transforms; Cambridge University Press. UK, 2009; pp. 1–858. [CrossRef]
- Rogers, S.; Girolami, M. A first course in machine learning, 2nd ed.; Chapman & Hall/CRC Press: Boca Raton, FL, USA, 2017; pp. 185–195. [Google Scholar]
- K-nearest neighbor. Available online: http://scholarpedia.org/article/K-nearest_neighbor (accessed on 16 August 2023).
- Mucherino, A., Papajorgji, P. J., Pardalos, P. M., Mucherino, A., Papajorgji, P. J., Pardalos, P. M. K-nearest neighbor classification. In Data mining in agriculture. Springer Optimization and Its Applications, 2nd ed.; Springer: New York, NY, USA, 2009; Volume 34, pp. 83–106. [Google Scholar] [CrossRef]
- Gerón, A. Hands-On Machine Learnign with Scikit-Learn, Keras & TensorFlow, 2nd ed.; O’Reily: Sebastopol, CA, USA, 2019; pp. 189–212. [Google Scholar]
- Breiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Ho, T. K. The random subspace method for constructing decision forests. IEEE Transactions on Pattern Analysis and Machine Intelligence 1998, 20, 832–844. [Google Scholar] [CrossRef]
- X. Ma, T. Yang, J. Chen and Z. Liu, k-Nearest Neighbor algorithm based on feature subspace, In Proceedings of the 2021 International Conference on Big Data Analysis and Computer Science (BDACS), Kunming, China, 25-27 June 2021. 27 June. [CrossRef]
- Xie, P., Zuo, K., Zhang, Y., Li, F., Yin, M., Lu, K. Interpretable classification from skin cancer histology slides using deep learning: A retrospective multicenter study. ArXiv 2019, preprint arXiv:1904.06156. arXiv:1904.06156. [CrossRef]
- Ogudo, K. A., Surendran, R., Khalaf, O. I. Optimal Artificial Intelligence Based Automated Skin Lesion Detection and Classification Model. Comput. Syst. Sci. Eng. 2023, 44, 693–707. [Google Scholar] [CrossRef]
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.997713 | 0.000435 | 0.992518 | 0.001918 | 0.998454 | 0.000441 | 0.989201 | 0.003089 |
| SCC | 0.994121 | 0.000502 | 0.989432 | 0.002688 | 0.994789 | 0.000394 | 0.96442 | 0.002662 |
| MEL | 0.995564 | 0.000634 | 0.978523 | 0.00349 | 0.997998 | 0.000466 | 0.985882 | 0.003276 |
| AK | 0.994141 | 0.00064 | 0.969387 | 0.004028 | 0.997692 | 0.000405 | 0.983691 | 0.002828 |
| BKL | 0.994707 | 0.000585 | 0.982108 | 0.003661 | 0.9965 | 0.000433 | 0.975644 | 0.002831 |
| DF | 0.996077 | 0.00053 | 0.975942 | 0.003758 | 0.998964 | 0.0003 | 0.992662 | 0.00211 |
| NV | 0.995768 | 0.000637 | 0.987118 | 0.003555 | 0.996996 | 0.00054 | 0.979067 | 0.003622 |
| VASC | 0.998452 | 0.000227 | 0.991186 | 0.001829 | 0.999489 | 0.000202 | 0.996405 | 0.001409 |
| MEAN±1SD | 0.995818 | 0.001585 | 0.983277 | 0.008205 | 0.99761 | 0.001502 | 0.983371 | 0.01024 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.998902 | 0.000771 | 0.995155 | 0.003451 | 0.999443 | 0.000747 | 0.996129 | 0.005223 |
| SCC | 0.996671 | 0.001348 | 0.994984 | 0.006641 | 0.99692 | 0.001477 | 0.978896 | 0.009978 |
| MEL | 0.997328 | 0.001484 | 0.984985 | 0.011026 | 0.999095 | 0.000836 | 0.993616 | 0.005861 |
| AK | 0.996467 | 0.001355 | 0.981774 | 0.008303 | 0.998527 | 0.001044 | 0.989513 | 0.007158 |
| BKL | 0.99692 | 0.001044 | 0.99105 | 0.006158 | 0.997775 | 0.001073 | 0.984501 | 0.00745 |
| DF | 0.997656 | 0.00109 | 0.984903 | 0.008138 | 0.999431 | 0.000583 | 0.995944 | 0.004108 |
| NV | 0.997464 | 0.001555 | 0.99428 | 0.006072 | 0.99791 | 0.00166 | 0.98595 | 0.010578 |
| VASC | 0.998913 | 0.000538 | 0.993741 | 0.004151 | 0.999651 | 0.000327 | 0.997529 | 0.002344 |
| MEAN±1SD | 0.99754 | 0.000933 | 0.990109 | 0.005392 | 0.998594 | 0.000981 | 0.99026 | 0.006679 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.997931 | 0.000475 | 0.991772 | 0.002502 | 0.998815 | 0.0004 | 0.99175 | 0.002773 |
| SCC | 0.998225 | 0.000515 | 0.9958 | 0.00246 | 0.998571 | 0.000366 | 0.990031 | 0.002564 |
| MEL | 0.998585 | 0.000376 | 0.995069 | 0.002159 | 0.999085 | 0.000309 | 0.993581 | 0.002148 |
| AK | 0.997857 | 0.000469 | 0.98977 | 0.002736 | 0.99901 | 0.000418 | 0.993046 | 0.002911 |
| BKL | 0.997951 | 0.000604 | 0.99258 | 0.003565 | 0.998719 | 0.000419 | 0.991067 | 0.002932 |
| DF | 0.998488 | 0.00042 | 0.993297 | 0.003042 | 0.99923 | 0.000292 | 0.994617 | 0.002015 |
| NV | 0.998321 | 0.000412 | 0.99141 | 0.002184 | 0.999311 | 0.000323 | 0.995173 | 0.002237 |
| VASC | 0.999768 | 0.000158 | 0.998817 | 0.001053 | 0.999903 | 0.000108 | 0.99932 | 0.000756 |
| MEAN±1SD | 0.998391 | 0.000617 | 0.993565 | 0.002882 | 0.99908 | 0.000417 | 0.993573 | 0.002906 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.999241 | 0.000701 | 0.997109 | 0.004113 | 0.999535 | 0.000714 | 0.996713 | 0.005081 |
| SCC | 0.999411 | 0.000667 | 0.998569 | 0.003459 | 0.999534 | 0.000685 | 0.996772 | 0.00483 |
| MEL | 0.999468 | 0.000548 | 0.998491 | 0.003087 | 0.999611 | 0.000568 | 0.997317 | 0.003941 |
| AK | 0.999207 | 0.000769 | 0.996019 | 0.004335 | 0.999665 | 0.000615 | 0.997598 | 0.004424 |
| BKL | 0.999343 | 0.000807 | 0.997474 | 0.005446 | 0.999612 | 0.000638 | 0.997301 | 0.004364 |
| DF | 0.999683 | 0.000527 | 0.999094 | 0.002063 | 0.999767 | 0.000507 | 0.998393 | 0.003469 |
| NV | 0.999162 | 0.000816 | 0.995287 | 0.00487 | 0.999716 | 0.000584 | 0.997986 | 0.004132 |
| VASC | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| MEAN±1SD | 0.99944 | 0.000281 | 0.997755 | 0.001587 | 0.99968 | 0.000153 | 0.99776 | 0.001067 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.990617 | 0.001202 | 0.938094 | 0.009789 | 0.998108 | 0.000634 | 0.986097 | 0.004562 |
| SCC | 0.991729 | 0.000776 | 0.9374 | 0.006256 | 0.999511 | 0.000248 | 0.996384 | 0.001816 |
| MEL | 0.990704 | 0.000929 | 0.948301 | 0.004959 | 0.99677 | 0.000679 | 0.976802 | 0.004607 |
| AK | 0.964331 | 0.002284 | 0.992258 | 0.002383 | 0.960355 | 0.002588 | 0.781032 | 0.01118 |
| BKL | 0.988485 | 0.000888 | 0.915523 | 0.006838 | 0.998884 | 0.000384 | 0.991531 | 0.002881 |
| DF | 0.993657 | 0.000814 | 0.963894 | 0.006035 | 0.997907 | 0.000437 | 0.985045 | 0.003063 |
| NV | 0.987007 | 0.000943 | 0.955562 | 0.006548 | 0.991504 | 0.000797 | 0.941491 | 0.00538 |
| VASC | 0.993679 | 0.000848 | 0.949736 | 0.006868 | 0.999951 | 7.96E-05 | 0.999643 | 0.000587 |
| MEAN±1SD | 0.987526 | 0.009651 | 0.950096 | 0.022366 | 0.992874 | 0.013403 | 0.957253 | 0.073477 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.994792 | 0.001981 | 0.963688 | 0.014052 | 0.999249 | 0.000668 | 0.994572 | 0.004833 |
| SCC | 0.996071 | 0.000904 | 0.968772 | 0.007678 | 0.999922 | 0.000214 | 0.999446 | 0.001521 |
| MEL | 0.996535 | 0.001641 | 0.980529 | 0.012471 | 0.998837 | 0.00096 | 0.991698 | 0.006806 |
| AK | 0.982756 | 0.002737 | 0.994899 | 0.00495 | 0.980997 | 0.002822 | 0.883549 | 0.015957 |
| BKL | 0.99435 | 0.001641 | 0.960839 | 0.010733 | 0.999222 | 0.000671 | 0.994451 | 0.004731 |
| DF | 0.997226 | 0.001441 | 0.983665 | 0.008772 | 0.999148 | 0.00091 | 0.993885 | 0.006639 |
| NV | 0.994124 | 0.001773 | 0.982219 | 0.008845 | 0.995812 | 0.001428 | 0.970819 | 0.010054 |
| VASC | 0.997271 | 0.001331 | 0.978214 | 0.010582 | 1 | 0 | 1 | 0 |
| MEAN±1SD | 0.994141 | 0.004764 | 0.976603 | 0.011407 | 0.996649 | 0.006459 | 0.978552 | 0.039459 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.970352 | 0.001239 | 0.904254 | 0.007983 | 0.979737 | 0.001104 | 0.863793 | 0.006639 |
| SCC | 0.973061 | 0.001097 | 0.893946 | 0.007351 | 0.984325 | 0.00116 | 0.890457 | 0.006616 |
| MEL | 0.970867 | 0.000961 | 0.829685 | 0.006684 | 0.991048 | 0.000499 | 0.929828 | 0.003744 |
| AK | 0.977499 | 0.000898 | 0.897616 | 0.006106 | 0.988921 | 0.00103 | 0.920629 | 0.006421 |
| BKL | 0.972529 | 0.000865 | 0.865436 | 0.00704 | 0.987814 | 0.000886 | 0.910286 | 0.005526 |
| DF | 0.985904 | 0.000616 | 0.921273 | 0.004687 | 0.995152 | 0.000607 | 0.96458 | 0.004144 |
| NV | 0.959978 | 0.000896 | 0.944947 | 0.003669 | 0.96213 | 0.001128 | 0.781547 | 0.004489 |
| VASC | 0.991525 | 0.000697 | 0.949302 | 0.005961 | 0.997563 | 0.000433 | 0.982402 | 0.002982 |
| MEAN±1SD | 0.975214 | 0.009798 | 0.900808 | 0.039817 | 0.985836 | 0.011125 | 0.90544 | 0.062767 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.977502 | 0.003381 | 0.93541 | 0.015774 | 0.98365 | 0.003091 | 0.89323 | 0.018677 |
| SCC | 0.979337 | 0.002892 | 0.921498 | 0.020366 | 0.987716 | 0.0027 | 0.915755 | 0.017457 |
| MEL | 0.976551 | 0.003395 | 0.858984 | 0.024565 | 0.993301 | 0.00136 | 0.947996 | 0.010845 |
| AK | 0.983243 | 0.002995 | 0.913926 | 0.013777 | 0.993147 | 0.002206 | 0.949931 | 0.015829 |
| BKL | 0.978023 | 0.003674 | 0.893106 | 0.022957 | 0.990187 | 0.001856 | 0.928795 | 0.012963 |
| DF | 0.991078 | 0.001802 | 0.947692 | 0.011963 | 0.997261 | 0.001098 | 0.979965 | 0.008013 |
| NV | 0.966044 | 0.004155 | 0.952837 | 0.010116 | 0.967897 | 0.005143 | 0.807599 | 0.024077 |
| VASC | 0.993671 | 0.001883 | 0.959445 | 0.013131 | 0.998526 | 0.000779 | 0.989305 | 0.005602 |
| MEAN±1SD | 0.980681 | 0.008731 | 0.922862 | 0.033929 | 0.988961 | 0.009796 | 0.926572 | 0.057544 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.996612 | 0.000647 | 0.988253 | 0.003377 | 0.997809 | 0.000452 | 0.984767 | 0.003143 |
| SCC | 0.996459 | 0.000687 | 0.992086 | 0.00247 | 0.997082 | 0.000661 | 0.979832 | 0.004407 |
| MEL | 0.991981 | 0.000865 | 0.967874 | 0.005457 | 0.995412 | 0.00063 | 0.967787 | 0.004308 |
| AK | 0.997019 | 0.000537 | 0.98884 | 0.00308 | 0.998188 | 0.000476 | 0.98736 | 0.003258 |
| BKL | 0.993662 | 0.000735 | 0.968304 | 0.005179 | 0.997277 | 0.000491 | 0.980674 | 0.003422 |
| DF | 0.995847 | 0.00054 | 0.984048 | 0.003433 | 0.997534 | 0.000475 | 0.9828 | 0.003276 |
| NV | 0.9906 | 0.000995 | 0.962863 | 0.005059 | 0.994565 | 0.000698 | 0.962032 | 0.004756 |
| VASC | 0.997852 | 0.000516 | 0.987744 | 0.003771 | 0.999295 | 0.000296 | 0.995033 | 0.002071 |
| MEAN±1SD | 0.995004 | 0.002616 | 0.980001 | 0.011626 | 0.997145 | 0.001511 | 0.980036 | 0.010578 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.998562 | 0.000776 | 0.995856 | 0.00456 | 0.998941 | 0.00065 | 0.992512 | 0.004646 |
| SCC | 0.998709 | 0.000954 | 0.997816 | 0.002984 | 0.998835 | 0.000948 | 0.991932 | 0.006699 |
| MEL | 0.996581 | 0.001444 | 0.98485 | 0.010675 | 0.998316 | 0.001431 | 0.988392 | 0.009868 |
| AK | 0.998913 | 0.001019 | 0.996475 | 0.005015 | 0.999275 | 0.000801 | 0.994956 | 0.005579 |
| BKL | 0.997101 | 0.001219 | 0.984999 | 0.007762 | 0.998836 | 0.001008 | 0.991779 | 0.007134 |
| DF | 0.998358 | 0.000953 | 0.995091 | 0.004975 | 0.998823 | 0.000948 | 0.991805 | 0.006562 |
| NV | 0.995811 | 0.001733 | 0.983651 | 0.008814 | 0.997541 | 0.001488 | 0.982872 | 0.010041 |
| VASC | 0.999026 | 0.000761 | 0.993904 | 0.006181 | 0.999754 | 0.00033 | 0.998266 | 0.002323 |
| MEAN±1SD | 0.997883 | 0.001215 | 0.99158 | 0.00598 | 0.99879 | 0.000652 | 0.991564 | 0.004523 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.968962 | 0.001612 | 0.902908 | 0.008807 | 0.978419 | 0.001602 | 0.857063 | 0.008559 |
| SCC | 0.964886 | 0.001417 | 0.886476 | 0.009336 | 0.976084 | 0.001643 | 0.841252 | 0.008924 |
| MEL | 0.956256 | 0.001899 | 0.817315 | 0.011059 | 0.976133 | 0.001703 | 0.830625 | 0.009523 |
| AK | 0.966205 | 0.001226 | 0.862392 | 0.009765 | 0.98104 | 0.001444 | 0.866847 | 0.008002 |
| BKL | 0.963505 | 0.00187 | 0.84941 | 0.00938 | 0.979755 | 0.002278 | 0.856939 | 0.013198 |
| DF | 0.969574 | 0.001417 | 0.849535 | 0.010333 | 0.986664 | 0.001288 | 0.900786 | 0.008433 |
| NV | 0.951404 | 0.001651 | 0.834493 | 0.010815 | 0.968077 | 0.001882 | 0.788632 | 0.009132 |
| VASC | 0.985493 | 0.001017 | 0.902211 | 0.007839 | 0.997421 | 0.000526 | 0.980456 | 0.003871 |
| MEAN±1SD | 0.965786 | 0.010118 | 0.863092 | 0.031504 | 0.980449 | 0.008639 | 0.865325 | 0.056466 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.977899 | 0.002878 | 0.946323 | 0.014022 | 0.982366 | 0.003188 | 0.884054 | 0.017584 |
| SCC | 0.975928 | 0.003423 | 0.906904 | 0.021586 | 0.985793 | 0.00251 | 0.901248 | 0.016761 |
| MEL | 0.965308 | 0.004586 | 0.879849 | 0.023875 | 0.977466 | 0.003912 | 0.847469 | 0.023878 |
| AK | 0.97474 | 0.003065 | 0.891863 | 0.016402 | 0.986582 | 0.002767 | 0.904245 | 0.020062 |
| BKL | 0.971558 | 0.00339 | 0.897429 | 0.017104 | 0.982317 | 0.002985 | 0.879933 | 0.018989 |
| DF | 0.977219 | 0.002996 | 0.877022 | 0.024321 | 0.991761 | 0.002146 | 0.939128 | 0.015253 |
| NV | 0.960462 | 0.004176 | 0.85456 | 0.025912 | 0.975699 | 0.004218 | 0.83514 | 0.02571 |
| VASC | 0.988055 | 0.00234 | 0.912879 | 0.018859 | 0.998694 | 0.000701 | 0.990039 | 0.005171 |
| MEAN±1SD | 0.973896 | 0.008384 | 0.895854 | 0.027499 | 0.985085 | 0.0075 | 0.897657 | 0.049623 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.96834 | 0.00108 | 0.851159 | 0.010441 | 0.985047 | 0.001385 | 0.890453 | 0.008299 |
| SCC | 0.963689 | 0.001617 | 0.849973 | 0.013445 | 0.979887 | 0.001611 | 0.857745 | 0.009328 |
| MEL | 0.954031 | 0.001684 | 0.798347 | 0.012674 | 0.976334 | 0.002385 | 0.828959 | 0.012168 |
| AK | 0.965667 | 0.001554 | 0.867501 | 0.009935 | 0.9797 | 0.002158 | 0.859622 | 0.011847 |
| BKL | 0.959197 | 0.001785 | 0.813381 | 0.012224 | 0.979997 | 0.001902 | 0.853189 | 0.010953 |
| DF | 0.969908 | 0.001379 | 0.833521 | 0.010051 | 0.989456 | 0.001226 | 0.918975 | 0.008551 |
| NV | 0.941463 | 0.002009 | 0.890112 | 0.013375 | 0.94879 | 0.00315 | 0.71319 | 0.010549 |
| VASC | 0.981986 | 0.001287 | 0.91312 | 0.008143 | 0.991796 | 0.001024 | 0.940725 | 0.006908 |
| MEAN±1SD | 0.963035 | 0.01197 | 0.852139 | 0.038069 | 0.978876 | 0.013264 | 0.857857 | 0.069131 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.977434 | 0.002263 | 0.911182 | 0.014176 | 0.986988 | 0.002337 | 0.909599 | 0.016076 |
| SCC | 0.972271 | 0.003538 | 0.871525 | 0.020069 | 0.986818 | 0.002835 | 0.904842 | 0.020734 |
| MEL | 0.9643 | 0.00416 | 0.853384 | 0.026301 | 0.979937 | 0.0029 | 0.857077 | 0.018503 |
| AK | 0.973607 | 0.003901 | 0.895428 | 0.016813 | 0.98474 | 0.003515 | 0.893324 | 0.023071 |
| BKL | 0.967969 | 0.004153 | 0.871239 | 0.02185 | 0.981898 | 0.003421 | 0.873844 | 0.020752 |
| DF | 0.974411 | 0.00296 | 0.85076 | 0.016279 | 0.99186 | 0.002333 | 0.936737 | 0.016802 |
| NV | 0.95608 | 0.003435 | 0.912351 | 0.018579 | 0.962345 | 0.004294 | 0.7758 | 0.022732 |
| VASC | 0.985847 | 0.00222 | 0.921673 | 0.014345 | 0.995114 | 0.001557 | 0.964687 | 0.010787 |
| MEAN±1SD | 0.97149 | 0.008917 | 0.885943 | 0.027834 | 0.983713 | 0.009942 | 0.889489 | 0.057024 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.967544 | 0.001553 | 0.892731 | 0.011109 | 0.978245 | 0.00162 | 0.85463 | 0.008652 |
| SCC | 0.958837 | 0.002056 | 0.87173 | 0.011594 | 0.971249 | 0.002404 | 0.812395 | 0.011305 |
| MEL | 0.951254 | 0.001966 | 0.792743 | 0.016617 | 0.973954 | 0.002412 | 0.813743 | 0.012396 |
| AK | 0.961603 | 0.001413 | 0.843246 | 0.008752 | 0.978504 | 0.001808 | 0.84871 | 0.010153 |
| BKL | 0.958948 | 0.001545 | 0.83026 | 0.009622 | 0.977347 | 0.001845 | 0.839881 | 0.010704 |
| DF | 0.965498 | 0.001278 | 0.833468 | 0.010531 | 0.984356 | 0.001613 | 0.884018 | 0.009842 |
| NV | 0.950286 | 0.001692 | 0.834216 | 0.013508 | 0.966907 | 0.002334 | 0.783326 | 0.010768 |
| VASC | 0.984078 | 0.000932 | 0.893935 | 0.007509 | 0.996871 | 0.000587 | 0.975965 | 0.004312 |
| MEAN±1SD | 0.962256 | 0.010704 | 0.849041 | 0.034759 | 0.978429 | 0.009109 | 0.851583 | 0.058924 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.977231 | 0.002806 | 0.950159 | 0.012809 | 0.98111 | 0.003381 | 0.876928 | 0.021674 |
| SCC | 0.970211 | 0.003376 | 0.883554 | 0.021992 | 0.982757 | 0.003779 | 0.880785 | 0.024607 |
| MEL | 0.962183 | 0.00427 | 0.867746 | 0.025005 | 0.975543 | 0.003754 | 0.83374 | 0.023414 |
| AK | 0.970652 | 0.003369 | 0.869158 | 0.015176 | 0.985198 | 0.003215 | 0.893751 | 0.021674 |
| BKL | 0.966769 | 0.003754 | 0.882164 | 0.019933 | 0.978868 | 0.003254 | 0.856108 | 0.020597 |
| DF | 0.972883 | 0.003797 | 0.859058 | 0.019839 | 0.989167 | 0.002575 | 0.919144 | 0.017968 |
| NV | 0.960417 | 0.003748 | 0.849669 | 0.027438 | 0.976155 | 0.004667 | 0.834863 | 0.027576 |
| VASC | 0.986458 | 0.002753 | 0.907463 | 0.018557 | 0.998054 | 0.000889 | 0.985479 | 0.006777 |
| MEAN±1SD | 0.97085 | 0.008363 | 0.883622 | 0.032105 | 0.983357 | 0.00748 | 0.8851 | 0.049853 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.99101 | 0.000853 | 0.966399 | 0.004479 | 0.99452 | 0.000799 | 0.961846 | 0.005073 |
| SCC | 0.990489 | 0.000854 | 0.973897 | 0.005359 | 0.992855 | 0.000691 | 0.951129 | 0.004412 |
| MEL | 0.98292 | 0.001179 | 0.929858 | 0.006006 | 0.990501 | 0.000993 | 0.933329 | 0.006406 |
| AK | 0.990648 | 0.000956 | 0.961649 | 0.00594 | 0.994779 | 0.000829 | 0.963376 | 0.005514 |
| BKL | 0.987704 | 0.001124 | 0.94218 | 0.007959 | 0.994208 | 0.000832 | 0.958831 | 0.005584 |
| DF | 0.991358 | 0.000866 | 0.961126 | 0.005819 | 0.995669 | 0.000647 | 0.969407 | 0.004409 |
| NV | 0.978637 | 0.001198 | 0.930944 | 0.008927 | 0.985455 | 0.001282 | 0.901657 | 0.006904 |
| VASC | 0.994882 | 0.000687 | 0.964408 | 0.004778 | 0.999236 | 0.000312 | 0.994494 | 0.002249 |
| MEAN±1SD | 0.988456 | 0.005248 | 0.953808 | 0.016992 | 0.993403 | 0.00405 | 0.954259 | 0.02732 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.995143 | 0.001416 | 0.980286 | 0.006849 | 0.997283 | 0.001247 | 0.980993 | 0.008724 |
| SCC | 0.995709 | 0.001617 | 0.989698 | 0.008021 | 0.996571 | 0.001178 | 0.976347 | 0.008376 |
| MEL | 0.989232 | 0.002362 | 0.953552 | 0.012095 | 0.994358 | 0.001925 | 0.960311 | 0.013101 |
| AK | 0.995822 | 0.001604 | 0.982016 | 0.008189 | 0.997812 | 0.001377 | 0.984743 | 0.009674 |
| BKL | 0.993795 | 0.002086 | 0.967452 | 0.010927 | 0.997569 | 0.001363 | 0.982643 | 0.009614 |
| DF | 0.995958 | 0.001499 | 0.984228 | 0.007814 | 0.997658 | 0.001094 | 0.983658 | 0.007641 |
| NV | 0.985892 | 0.002686 | 0.957146 | 0.013847 | 0.989976 | 0.00227 | 0.931492 | 0.013995 |
| VASC | 0.997543 | 0.001159 | 0.982462 | 0.007462 | 0.99969 | 0.000459 | 0.997759 | 0.003378 |
| MEAN±1SD | 0.993637 | 0.003989 | 0.974605 | 0.013462 | 0.996365 | 0.002976 | 0.974743 | 0.020327 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.998053 | 0.000557 | 0.991325 | 0.003305 | 0.999013 | 0.000478 | 0.993097 | 0.003319 |
| SCC | 0.996912 | 0.000734 | 0.985676 | 0.004545 | 0.998515 | 0.000531 | 0.989579 | 0.003716 |
| MEL | 0.997727 | 0.000607 | 0.990838 | 0.002973 | 0.998709 | 0.000528 | 0.990981 | 0.003639 |
| AK | 0.996932 | 0.000557 | 0.991146 | 0.00306 | 0.997758 | 0.00052 | 0.984448 | 0.003544 |
| BKL | 0.995689 | 0.00073 | 0.98531 | 0.004189 | 0.997167 | 0.00062 | 0.980236 | 0.004258 |
| DF | 0.997554 | 0.000714 | 0.987148 | 0.003966 | 0.999039 | 0.000378 | 0.993235 | 0.002618 |
| NV | 0.997815 | 0.000605 | 0.991293 | 0.003389 | 0.998745 | 0.000487 | 0.991221 | 0.003352 |
| VASC | 0.99912 | 0.000359 | 0.996388 | 0.001829 | 0.999511 | 0.000323 | 0.996589 | 0.00225 |
| MEAN±1SD | 0.997475 | 0.001002 | 0.98989 | 0.003684 | 0.998557 | 0.000754 | 0.989923 | 0.00524 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.999241 | 0.000712 | 0.996774 | 0.003976 | 0.999586 | 0.000548 | 0.997078 | 0.00388 |
| SCC | 0.998992 | 0.001031 | 0.994303 | 0.007762 | 0.999665 | 0.000483 | 0.997583 | 0.003562 |
| MEL | 0.999479 | 0.000826 | 0.998582 | 0.002601 | 0.999611 | 0.000737 | 0.997366 | 0.004978 |
| AK | 0.999253 | 0.000693 | 0.997617 | 0.003347 | 0.999483 | 0.00057 | 0.996359 | 0.003996 |
| BKL | 0.998471 | 0.001062 | 0.996698 | 0.003667 | 0.998732 | 0.001071 | 0.991193 | 0.00753 |
| DF | 0.999377 | 0.000606 | 0.99615 | 0.003906 | 0.999844 | 0.000283 | 0.998946 | 0.001909 |
| NV | 0.999298 | 0.000719 | 0.997054 | 0.004332 | 0.999625 | 0.000591 | 0.997368 | 0.004074 |
| VASC | 0.999841 | 0.000365 | 0.998721 | 0.002971 | 1 | 0 | 1 | 0 |
| MEAN±1SD | 0.999244 | 0.000395 | 0.996987 | 0.001413 | 0.999568 | 0.000375 | 0.996987 | 0.002606 |
| TRAIN SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.999462 | 0.000296 | 0.997958 | 0.001851 | 0.999677 | 0.000245 | 0.997737 | 0.0017 |
| SCC | 0.999038 | 0.000615 | 0.995829 | 0.002757 | 0.999496 | 0.000445 | 0.99646 | 0.003122 |
| MEL | 0.998709 | 0.000469 | 0.995324 | 0.003062 | 0.999194 | 0.000339 | 0.994398 | 0.002311 |
| AK | 0.998958 | 0.000546 | 0.995273 | 0.003405 | 0.999485 | 0.000285 | 0.996408 | 0.00199 |
| BKL | 0.999128 | 0.000355 | 0.997912 | 0.001905 | 0.999301 | 0.00039 | 0.995139 | 0.0027 |
| DF | 0.999176 | 0.000554 | 0.99656 | 0.002521 | 0.99955 | 0.000403 | 0.996853 | 0.002832 |
| NV | 0.998814 | 0.000494 | 0.99408 | 0.003025 | 0.999486 | 0.000315 | 0.996381 | 0.002206 |
| VASC | 0.999853 | 0.00013 | 0.999591 | 0.000708 | 0.99989 | 0.000134 | 0.999234 | 0.000934 |
| MEAN±1SD | 0.999142 | 0.000368 | 0.996566 | 0.001806 | 0.99951 | 0.000213 | 0.996576 | 0.001482 |
| TEST SET | ||||||||
| ACCURACY | SENSITIVITY | SPECIFICITY | PRECISION | |||||
| LESION | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD | MEAN | ±1SD |
| BCC | 0.999955 | 0.000248 | 1 | 0 | 0.999948 | 0.000284 | 0.999642 | 0.001963 |
| SCC | 0.999774 | 0.000515 | 0.998957 | 0.003181 | 0.999896 | 0.000394 | 0.999284 | 0.002726 |
| MEL | 0.999909 | 0.000345 | 1 | 0 | 0.999897 | 0.000393 | 0.999268 | 0.002791 |
| AK | 0.999909 | 0.000345 | 0.999643 | 0.001958 | 0.999948 | 0.000283 | 0.999637 | 0.00199 |
| BKL | 0.999864 | 0.000415 | 1 | 0 | 0.999845 | 0.000473 | 0.998915 | 0.003311 |
| DF | 0.999864 | 0.000415 | 0.998952 | 0.0032 | 1 | 0 | 1 | 0 |
| NV | 0.999909 | 0.000345 | 0.999282 | 0.002732 | 1 | 0 | 1 | 0 |
| VASC | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| MEAN±1SD | 0.999898 | 6.74E-05 | 0.999604 | 0.000475 | 0.999942 | 5.82E-05 | 0.999593 | 0.000407 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





