Submitted:
28 August 2023
Posted:
29 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Strain
2.2. Growth curve and determination of residual sugar content
2.3. Construction of transcriptome and processing of biological information
2.3.1. Material Handling
2.3.2. RNA extraction
2.3.3. Library Construction and Sequencing
2.3.4. Bioinformatics Analysis
3. Results
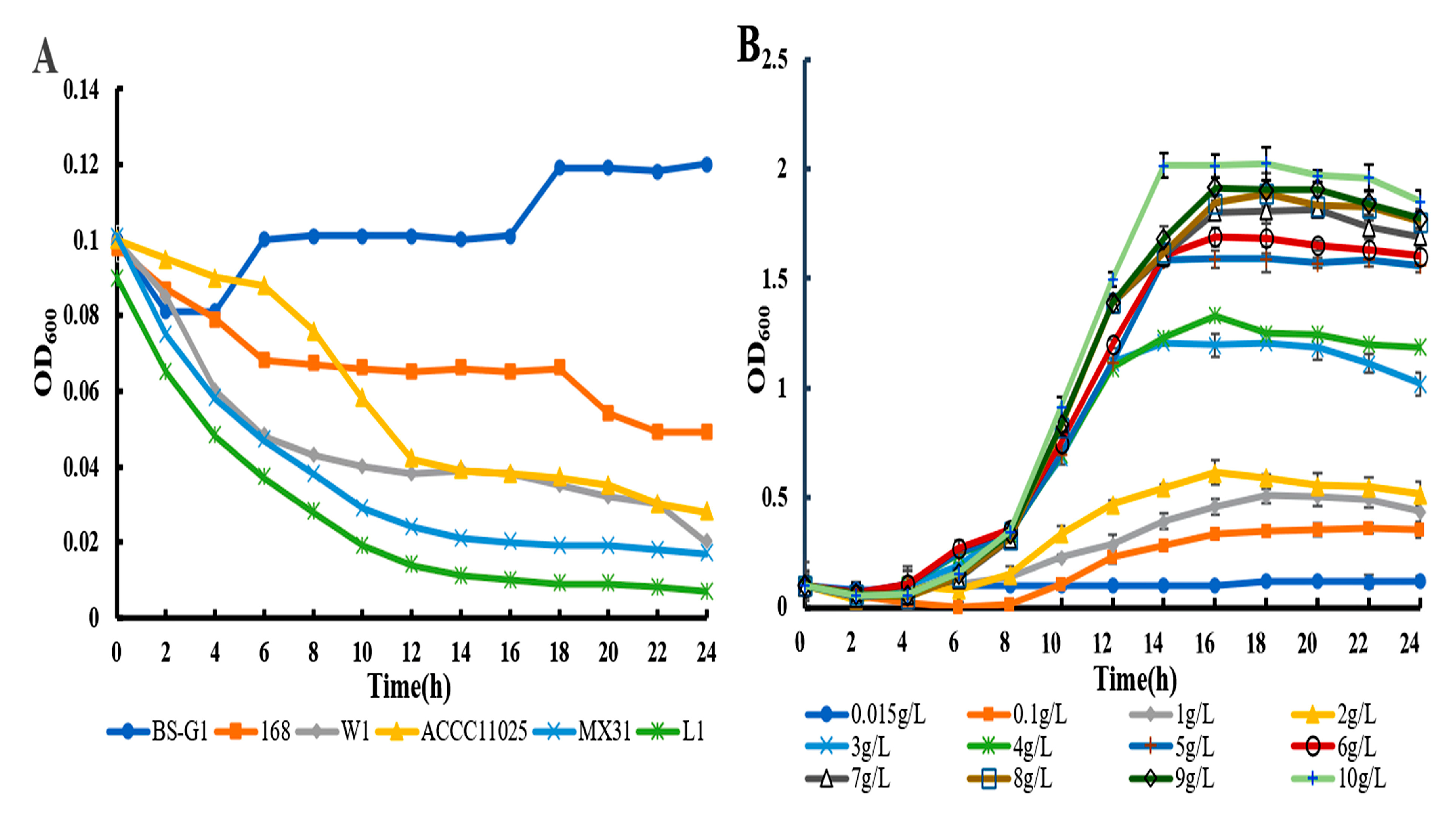
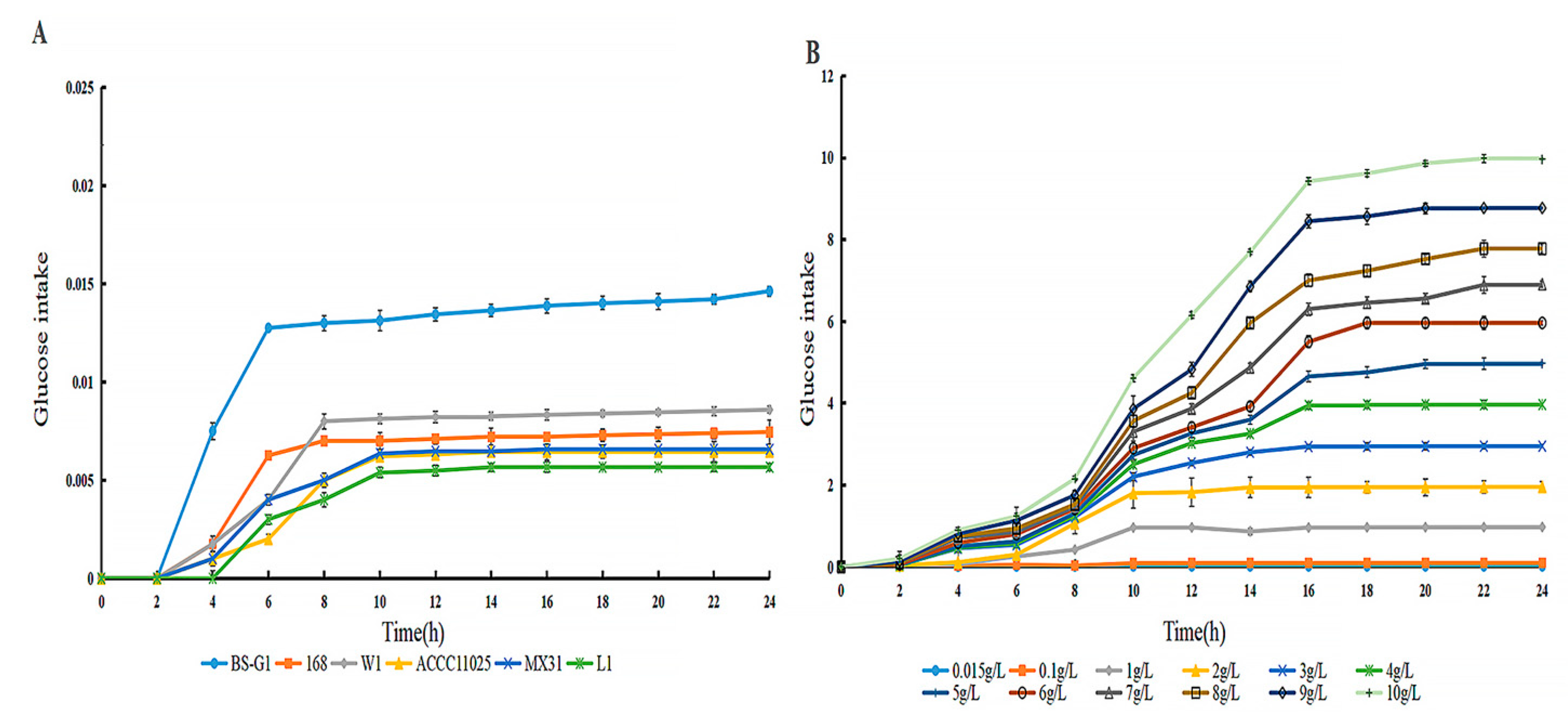
3.1. Growth Curves and Glucose Utilization
3.2. Illumina sequencing, quality filtering, and sequence alignment
3.3. Functional annotation and expression analysis of transcripts
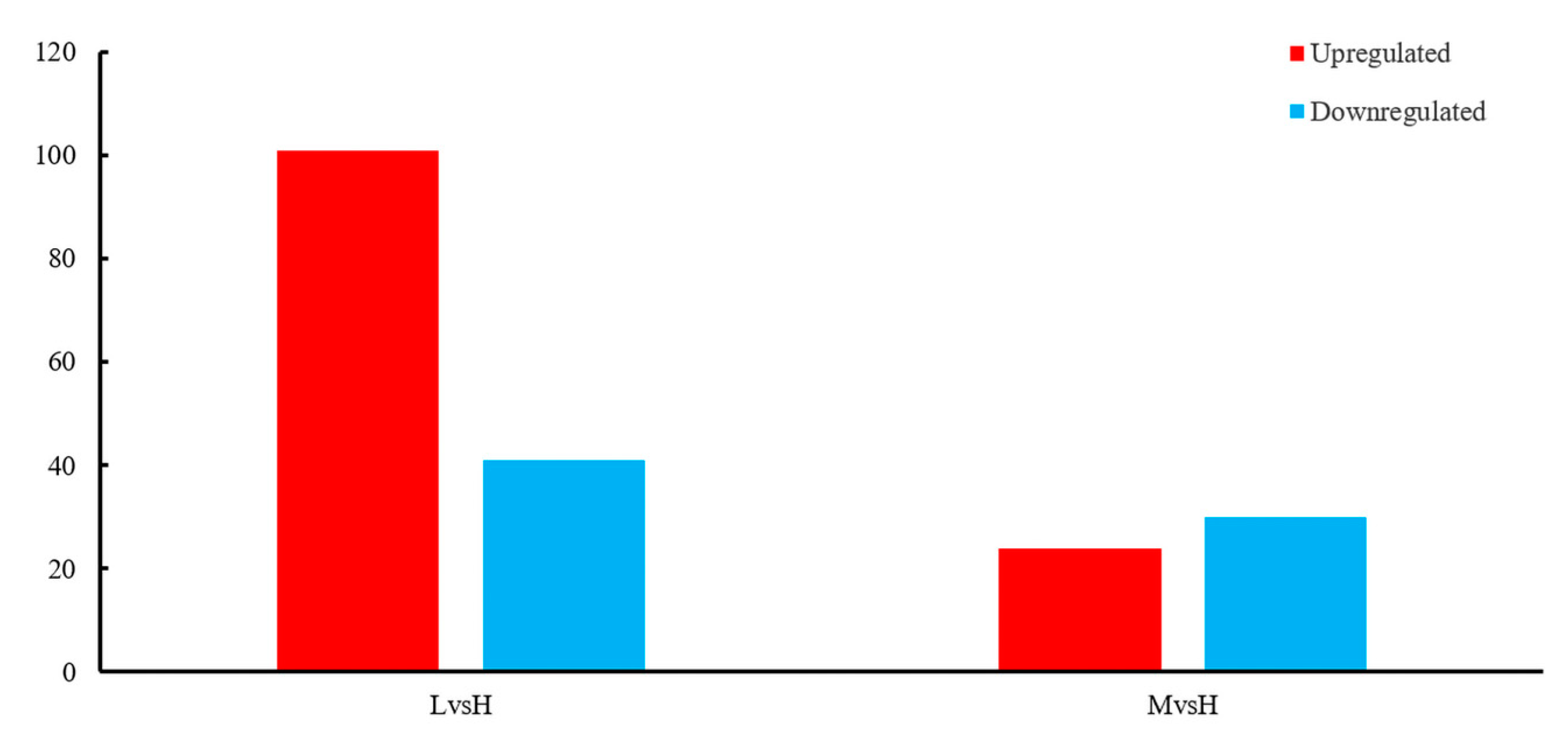
3.3. Identification of DEGs
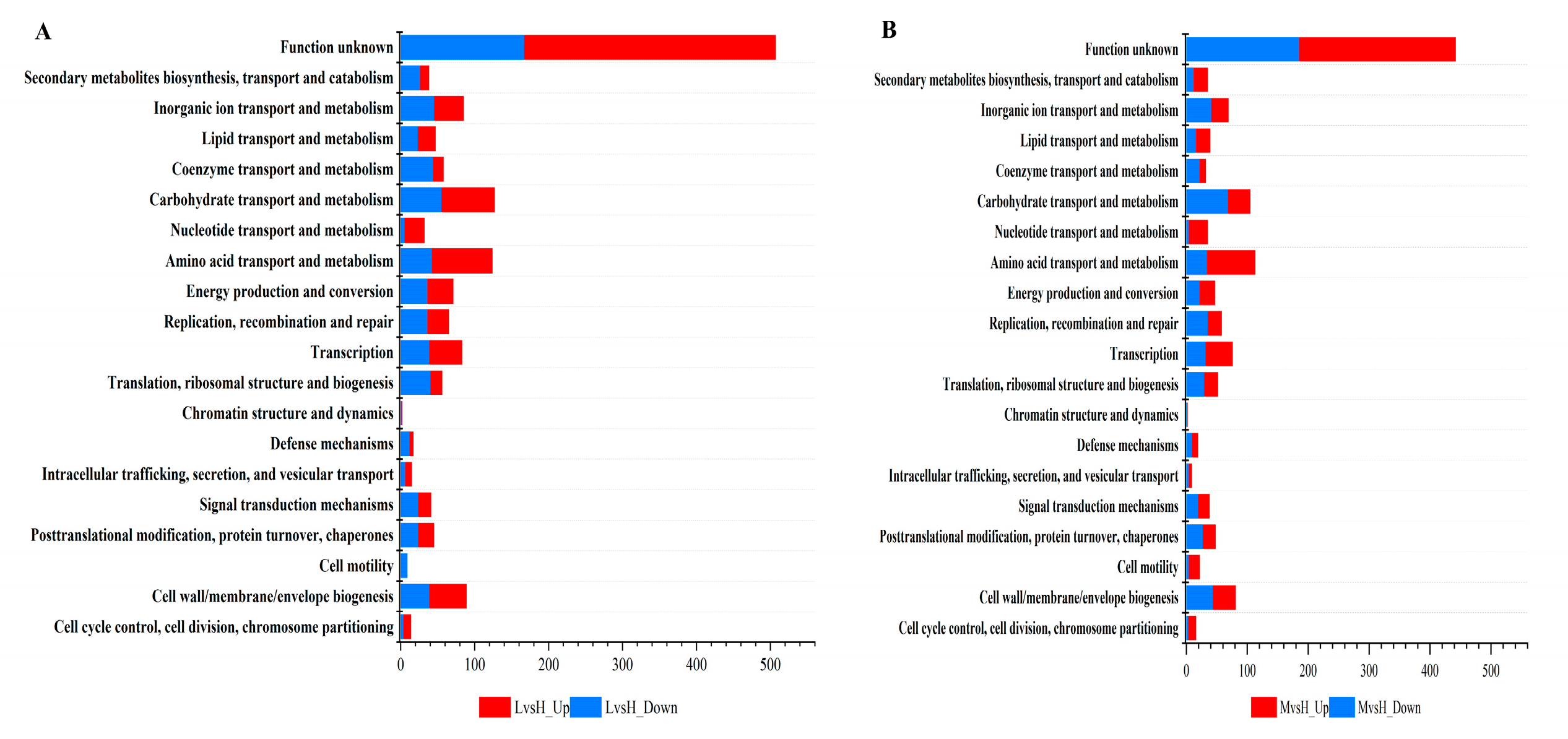
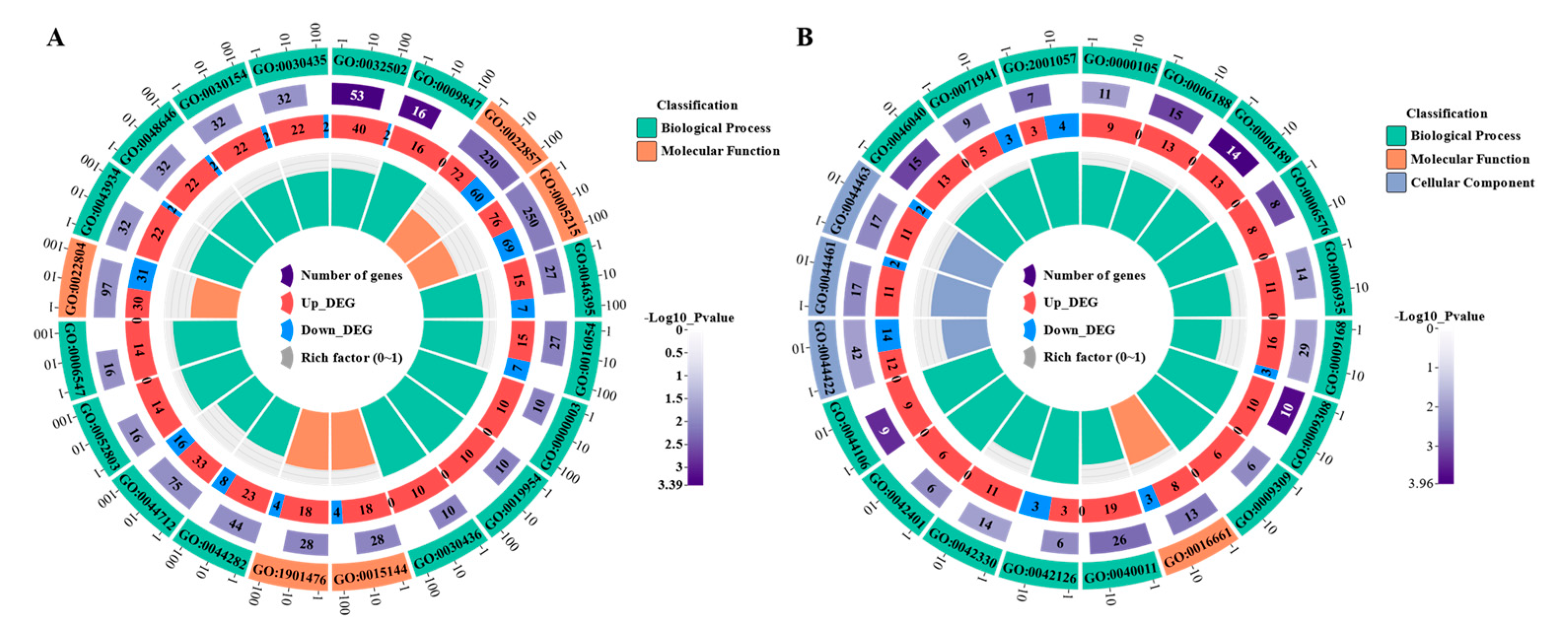
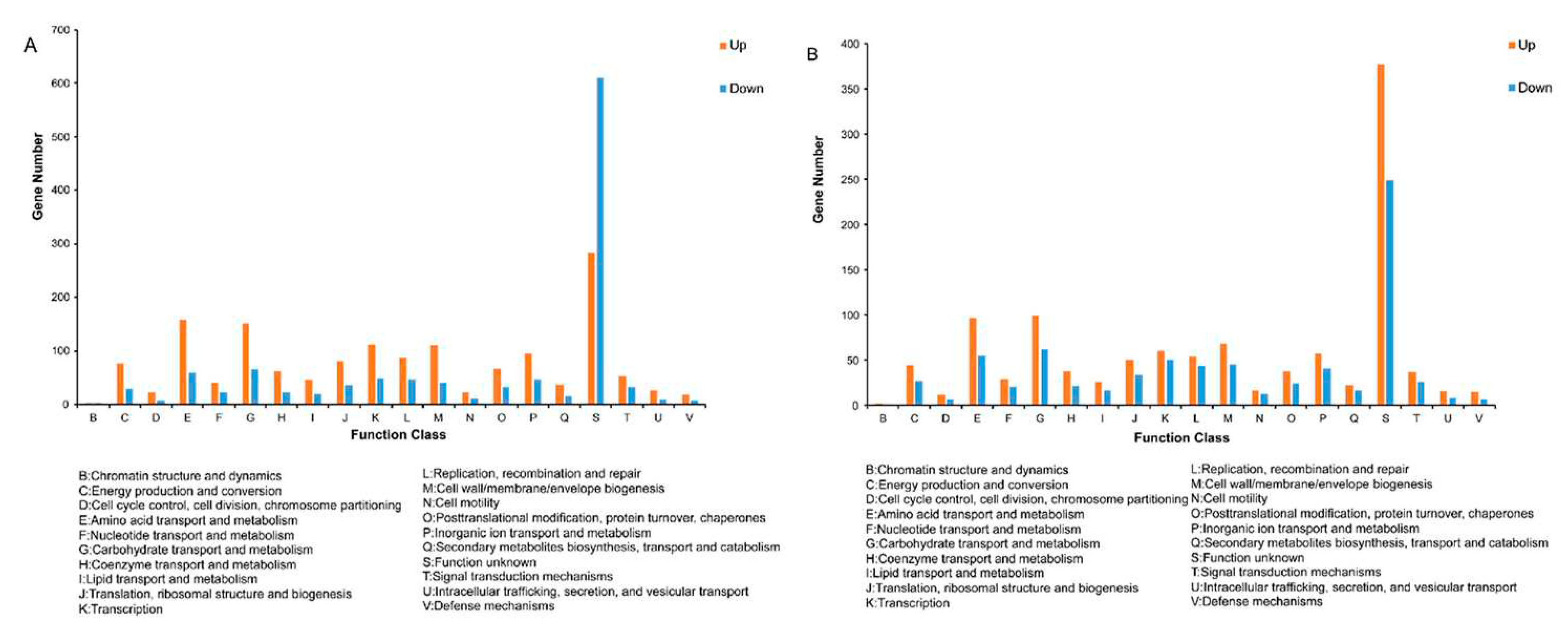
3.4. Functional annotation analysis of differentially expressed genes
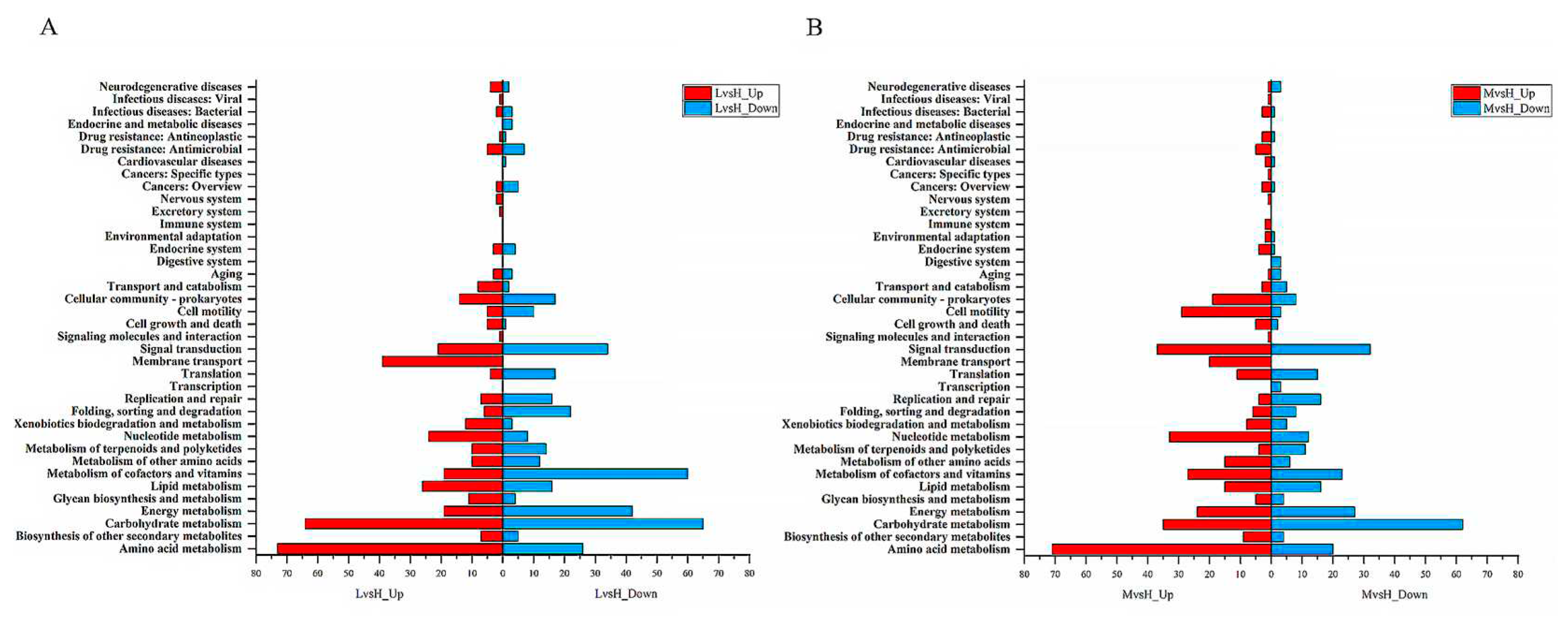
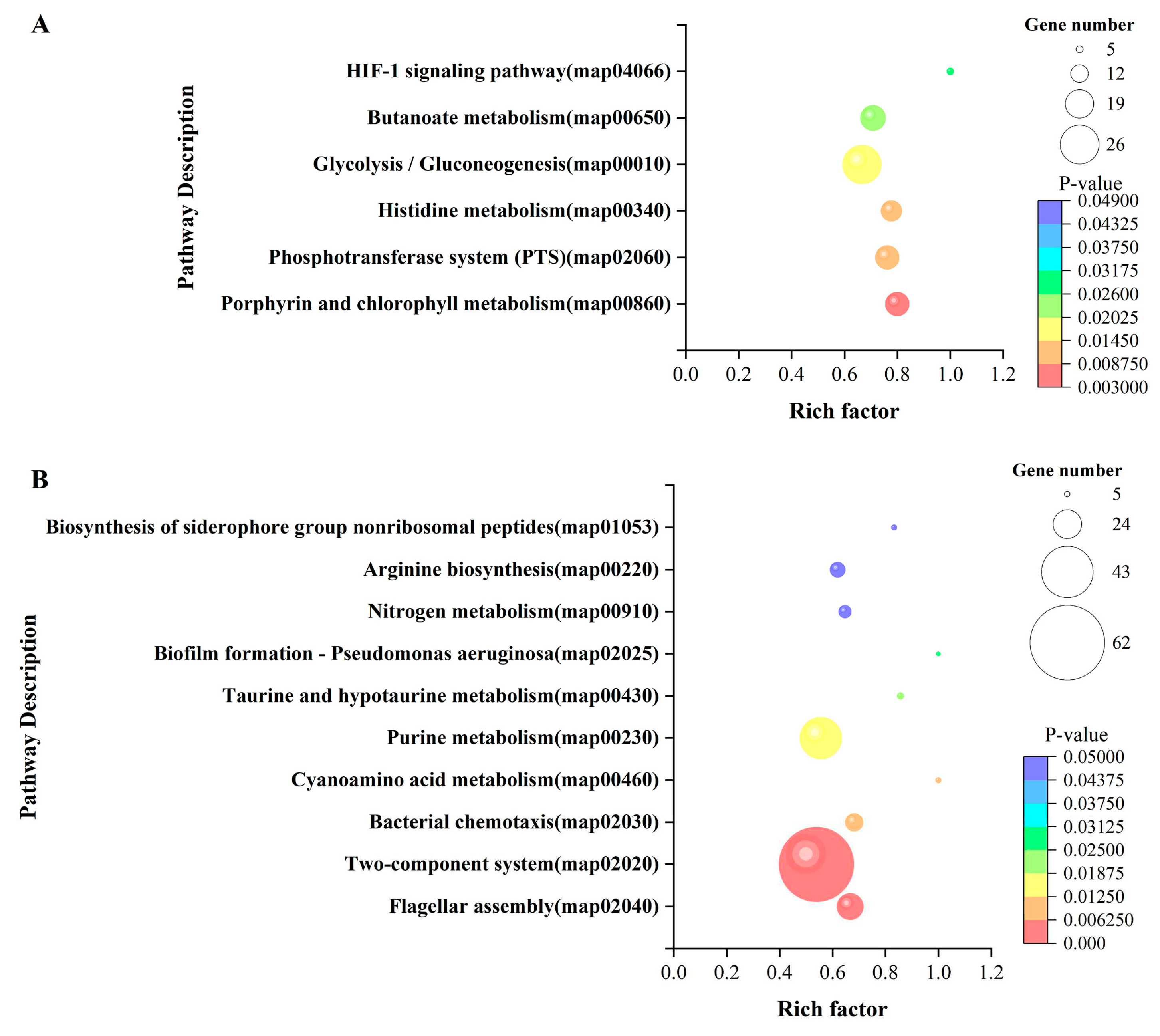
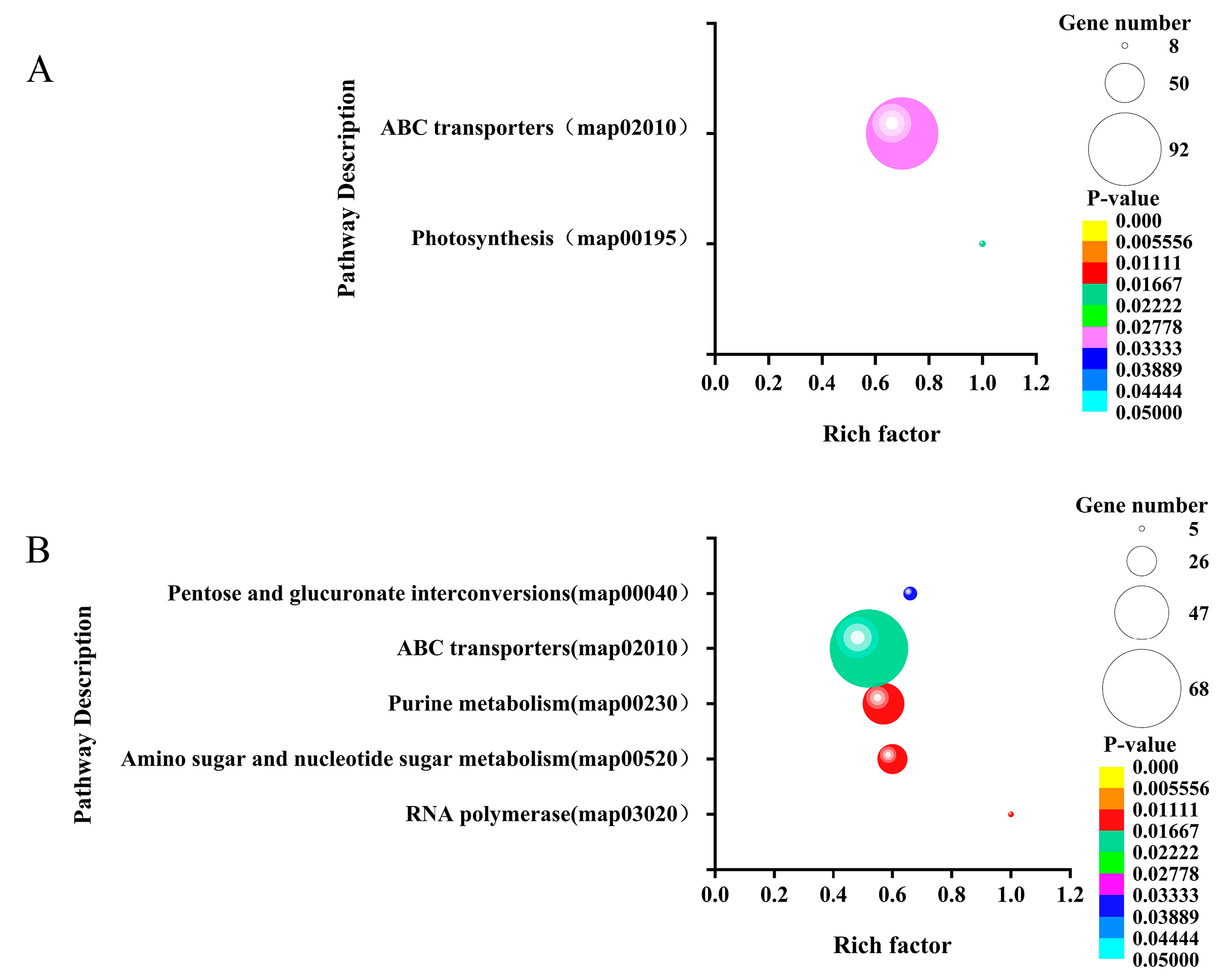
3.5. GO functional enrichment and KEGG pathway enrichment
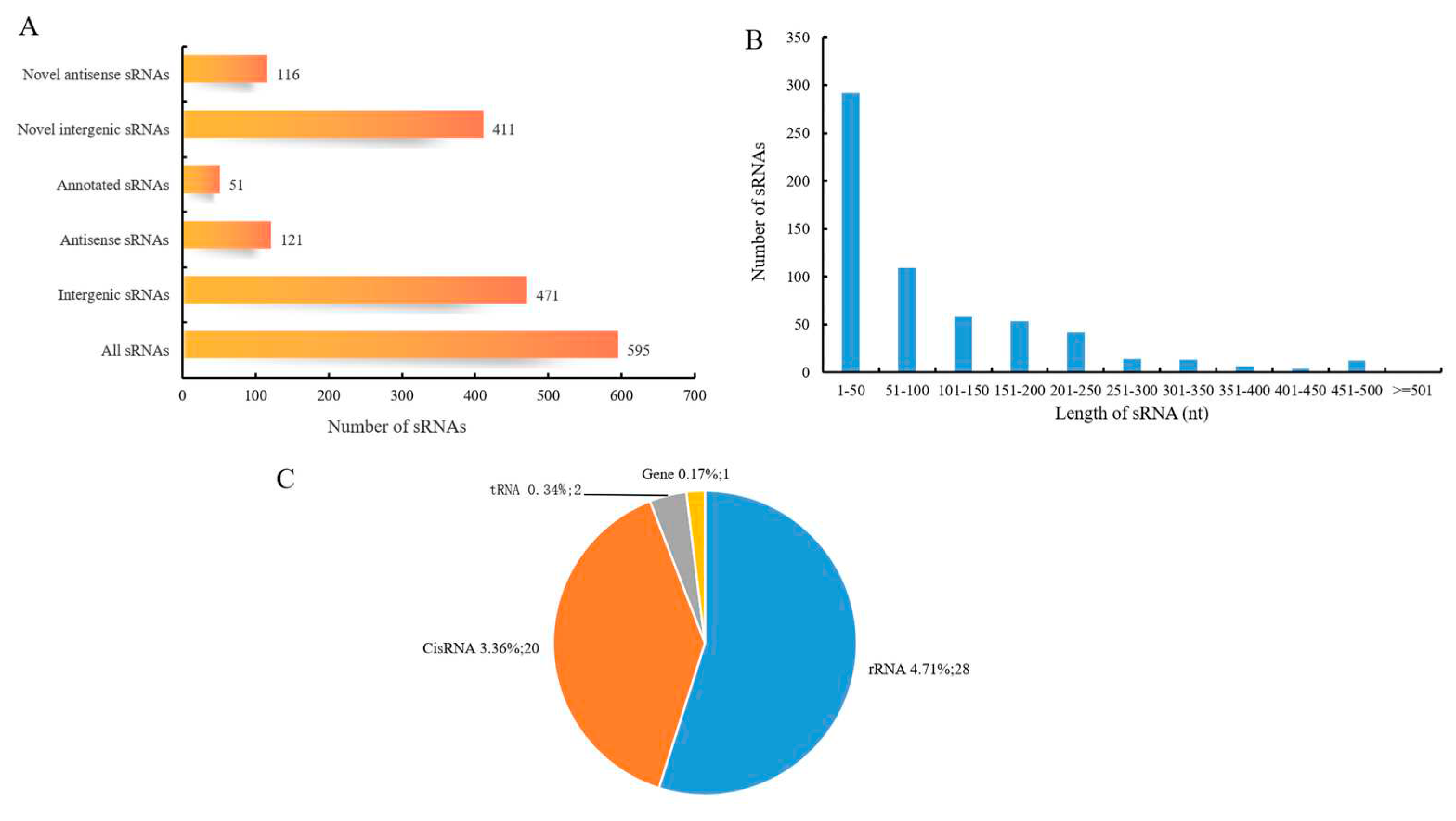
3.6. Identification of sRNA
3.7. Predict target genes of differentially expressed sRNAs
3.8. GO/KEGG enrichment analysis of sRNA target genes.
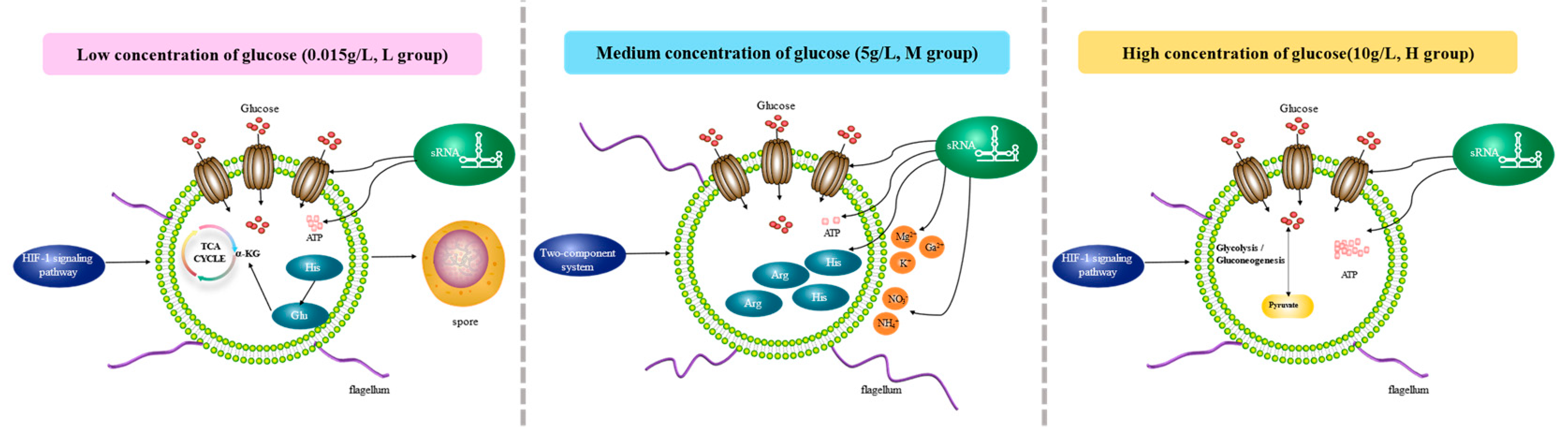
3.9. Adaptation patterns and key genes in a low-glucose environment
3.10. Critical genes for low-sugar adaptation.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Props, R.; Monsieurs, P.; Vandamme, P.; Leys, N.; Denef, V.J.; Boon, N. Gene Expansion and Positive Selection as Bacterial Adaptations to Oligotrophic Conditions. mSphere 2019, 4, e00011-19. [CrossRef]
- Chiriac, M.C.; Haber, M.; Salcher, M.M. Adaptive Genetic Traits in Pelagic Freshwater Microbes. Environ Microbiol 2023, 25, 606–641. [CrossRef]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberán, A. Life-History Strategies of Soil Microbial Communities in an Arid Ecosystem. ISME J 2021, 15, 649–657. [CrossRef]
- Cao, J.; Jiao, Y.; Che, R.; Holden, N.M.; Zhang, X.; Biswas, A.; Feng, Q. The Effects of Grazer Exclosure Duration on Soil Microbial Communities on the Qinghai-Tibetan Plateau. Sci Total Environ 2022, 839, 156238. [CrossRef]
- Dong, C.; Wei, L.; Wang, J.; Lai, Q.; Huang, Z.; Shao, Z. Genome-Based Taxonomic Rearrangement of Oceanobacter-Related Bacteria Including the Description of Thalassolituus Hydrocarbonoclasticus Sp. Nov. and Thalassolituus Pacificus Sp. Nov. and Emended Description of the Genus Thalassolituus. Front Microbiol 2022, 13, 1051202. [CrossRef]
- Ishida, Y.; Kadota, H. Growth Patterns and Substrate Requirements of Naturally Occurring Obligate Oligotrophs. Microb Ecol 1981, 7, 123–130. [CrossRef]
- de Araújo, H.L.; Martins, B.P.; Vicente, A.M.; Lorenzetti, A.P.R.; Koide, T.; Marques, M.V. Cold Regulation of Genes Encoding Ion Transport Systems in the Oligotrophic Bacterium Caulobacter Crescentus. Microbiol Spectr 2021, 9. [CrossRef]
- Ishida, Y.; Imai, I.; Miyagaki, T.; Kadota, H. Growth and Uptake Kinetics of a Facultatively Oligotrophic Bacterium at Low Nutrient Concentrations. Microb Ecol 1982, 8, 23–32. [CrossRef]
- Ogura, M.; Sato, T.; Abe, K. Bacillus Subtilis YlxR, Which Is Involved in Glucose-Responsive Metabolic Changes, Regulates Expression of TsaD for Protein Quality Control of Pyruvate Dehydrogenase. Frontiers in Microbiology 2019, 10, 1–15. [CrossRef]
- Park, Y.S.; Kai, K.; Iijima, S.; Kobayashi, T. Enhanced Beta-Galactosidase Production by High Cell-Density Culture of Recombinant Bacillus Subtilis with Glucose Concentration Control. Biotechnology & Bioengineering 1992, 40, 686. [CrossRef]
- Dedysh, S.N. Cultivating Uncultured Bacteria from Northern Wetlands: Knowledge Gained and Remaining Gaps. Front Microbiol 2011, 2, 184. [CrossRef]
- Staley, J.T. Budding Bacteria of the Pasteuria-Blastobacter Group. Can J Microbiol 1973, 19, 609–614. [CrossRef]
- Strous, M.; Pelletier, E.; Mangenot, S.; Rattei, T.; Lehner, A.; Taylor, M.W.; Horn, M.; Daims, H.; Bartol-Mavel, D.; Wincker, P.; et al. Deciphering the Evolution and Metabolism of an Anammox Bacterium from a Community Genome. Nature 2006, 440, 790–794. [CrossRef]
- Kaboré, O.D.; Godreuil, S.; Drancourt, M. Planctomycetes as Host-Associated Bacteria: A Perspective That Holds Promise for Their Future Isolations, by Mimicking Their Native Environmental Niches in Clinical Microbiology Laboratories. Front Cell Infect Microbiol 2020, 10, 519301. [CrossRef]
- Zhuang, H.; Wu, Z.; Xu, L.; Leu, S.-Y.; Lee, P.-H. Energy-Efficient Single-Stage Nitrite Shunt Denitrification with Saline Sewage through Concise Dissolved Oxygen (DO) Supply: Process Performance and Microbial Communities. Microorganisms 2020, 8, 919. [CrossRef]
- Koza, N.A.; Adedayo, A.A.; Babalola, O.O.; Kappo, A.P. Microorganisms in Plant Growth and Development: Roles in Abiotic Stress Tolerance and Secondary Metabolites Secretion. Microorganisms 2022, 10, 1528. [CrossRef]
- Carreón-Rodríguez, O.E.; Gosset, G.; Escalante, A.; Bolívar, F. Glucose Transport in Escherichia Coli: From Basics to Transport Engineering. Microorganisms 2023, 11, 1588. [CrossRef]
- Wen, J.; Zhao, X.; Si, F.; Qi, G. Surfactin, a Quorum Sensing Signal Molecule, Globally Affects the Carbon Metabolism in Bacillus Amyloliquefaciens. Metab Eng Commun 2021, 12, e00174. [CrossRef]
- Chamoli, S.; Kumar, P.; Navani, N.K.; Verma, A.K. Secretory Expression, Characterization and Docking Study of Glucose-Tolerant β-Glucosidase from B. Subtilis. International Journal of Biological Macromolecules 2016, 85, 425–433. [CrossRef]
- Pan, S.; Chen, G.; Wu, R.; Cao, X.; Liang, Z. Non-Sterile Submerged Fermentation of Fibrinolytic Enzyme by Marine Bacillus Subtilis Harboring Antibacterial Activity With Starvation Strategy. Frontiers in Microbiology 2019, 10, 1025. [CrossRef]
- Norris, N.; Levine, N.M.; Fernandez, V.I.; Stocker, R. Mechanistic Model of Nutrient Uptake Explains Dichotomy between Marine Oligotrophic and Copiotrophic Bacteria. PLoS Comput Biol 2021, 17, e1009023. [CrossRef]
- Wang, Y.; Hammes, F.; Boon, N.; Egli, T. Quantification of the Filterability of Freshwater Bacteria through 0.45, 0.22, and 0.1 Microm Pore Size Filters and Shape-Dependent Enrichment of Filterable Bacterial Communities. Environ Sci Technol 2007, 41, 7080–7086. [CrossRef]
- Sánchez-Cañizares, C.; Prell, J.; Pini, F.; Rutten, P.; Kraxner, K.; Wynands, B.; Karunakaran, R.; Poole, P.S. Global Control of Bacterial Nitrogen and Carbon Metabolism by a PTSNtr-Regulated Switch. Proc Natl Acad Sci U S A 2020, 117, 10234–10245. [CrossRef]
- Jeckelmann, J.M.; Erni, B. Carbohydrate Transport by Group Translocation: The Bacterial Phosphoenolpyruvate: Sugar Phosphotransferase System. Subcell Biochem 2019, 92, 223–274. [CrossRef]
- Fragoso-Jiménez, J.C.; Baert, J.; Nguyen, T.M.; Liu, W.; Sassi, H.; Goormaghtigh, F.; Van Melderen, L.; Gaytán, P.; Hernández-Chávez, G.; Martinez, A.; et al. Growth-Dependent Recombinant Product Formation Kinetics Can Be Reproduced through Engineering of Glucose Transport and Is Prone to Phenotypic Heterogeneity. Microb Cell Fact 2019, 18, 26. [CrossRef]
- Morabbi Heravi, K.; Altenbuchner, J. Cross Talk among Transporters of the Phosphoenolpyruvate-Dependent Phosphotransferase System in Bacillus Subtilis. J Bacteriol 2018, 200, e00213-18. [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell 2011, 43, 880–891. [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv Genet 2015, 90, 133–208. [CrossRef]
- Papenfort, K.; Melamed, S. Small RNAs, Large Networks: Posttranscriptional Regulons in Gram-Negative Bacteria. Annu Rev Microbiol 2023, 77, 36944261. [CrossRef]
- Taneja, S.; Dutta, T. On a Stake-out: Mycobacterial Small RNA Identification and Regulation. Noncoding RNA Res 2019, 4, 86–95. [CrossRef]
- Charbonnier, M.; González-Espinoza, G.; Kehl-Fie, T.E.; Lalaouna, D. Battle for Metals: Regulatory RNAs at the Front Line. Front Cell Infect Microbiol 2022, 12, 952948. [CrossRef]
- Parise, M.T.D.; Parise, D.; Aburjaile, F.F.; Pinto Gomide, A.C.; Kato, R.B.; Raden, M.; Backofen, R.; Azevedo, V.A. de C.; Baumbach, J. An Integrated Database of Small RNAs and Their Interplay With Transcriptional Gene Regulatory Networks in Corynebacteria. Front Microbiol 2021, 12, 656435. [CrossRef]
- Gottesman, S.; Storz, G. Bacterial Small RNA Regulators: Versatile Roles and Rapidly Evolving Variations. Cold Spring Harb Perspect Biol 2011, 3, a003798. [CrossRef]
- Fröhlich, K.S.; Papenfort, K. Regulation Outside the Box: New Mechanisms for Small RNAs. Mol Microbiol 2020, 114, 363–366. [CrossRef]
- Romilly, C.; Deindl, S.; Wagner, E.G.H. The Ribosomal Protein S1-Dependent Standby Site in TisB MRNA Consists of a Single-Stranded Region and a 5′ Structure Element. Proc Natl Acad Sci U S A 2019, 116, 15901–15906. [CrossRef]
- Melson, E.M.; Kendall, M.M. The SRNA DicF Integrates Oxygen Sensing to Enhance Enterohemorrhagic Escherichia Coli Virulence via Distinctive RNA Control Mechanisms. Proc Natl Acad Sci U S A 2019, 116, 14210–14215. [CrossRef]
- Balasubramanian, D.; Vanderpool, C.K. New Developments in Post-Transcriptional Regulation of Operons by Small RNAs. RNA Biol 2013, 10, 337–341. [CrossRef]
- Sheehan, L.M.; Budnick, J.A.; Fyffe-Blair, J.; King, K.A.; Settlage, R.E.; Caswell, C.C. The Endoribonuclease RNase E Coordinates Expression of MRNAs and Small Regulatory RNAs and Is Critical for the Virulence of Brucella Abortus. J Bacteriol 2020, 202, e00240-20. [CrossRef]
- Quendera, A.P.; Seixas, A.F.; dos Santos, R.F.; Santos, I.; Silva, J.P.N.; Arraiano, C.M.; Andrade, J.M. RNA-Binding Proteins Driving the Regulatory Activity of Small Non-Coding RNAs in Bacteria. Front Mol Biosci 2020, 7, 78. [CrossRef]
- McQuail, J.; Carpousis, A.J.; Wigneshweraraj, S. The Association between Hfq and RNase E in Long-Term Nitrogen-Starved Escherichia Coli. Mol Microbiol 2022, 117, 54–66. [CrossRef]
- De Lay, N.; Schu, D.J.; Gottesman, S. Bacterial Small RNA-Based Negative Regulation: Hfq and Its Accomplices. J Biol Chem 2013, 288, 7996–8003. [CrossRef]
- Murina, V.N.; Nikulin, A.D. Bacterial Small Regulatory RNAs and Hfq Protein. Biochemistry (Mosc) 2015, 80, 1647–1654. [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [CrossRef]
- Melamed, S.; Peer, A.; Faigenbaum-Romm, R.; Gatt, Y.E.; Reiss, N.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol Cell 2016, 63, 884–897. [CrossRef]
- Bobrovskyy, M.; Vanderpool, C.K. Regulation of Bacterial Metabolism by Small RNAs Using Diverse Mechanisms. Annu Rev Genet 2013, 47, 209–232. [CrossRef]
- Ford, B.A.; Sullivan, G.J.; Moore, L.; Varkey, D.; Zhu, H.; Ostrowski, M.; Mabbutt, B.C.; Paulsen, I.T.; Shah, B.S. Functional Characterisation of Substrate-Binding Proteins to Address Nutrient Uptake in Marine Picocyanobacteria. Biochem Soc Trans 2021, 49, 2465–2481. [CrossRef]
- Lauro, F.M.; McDougald, D.; Thomas, T.; Williams, T.J.; Egan, S.; Rice, S.; DeMaere, M.Z.; Ting, L.; Ertan, H.; Johnson, J.; et al. The Genomic Basis of Trophic Strategy in Marine Bacteria. Proc Natl Acad Sci U S A 2009, 106, 15527–15533. [CrossRef]
- Schwalbach, M.S.; Tripp, H.J.; Steindler, L.; Smith, D.P.; Giovannoni, S.J. The Presence of the Glycolysis Operon in SAR11 Genomes Is Positively Correlated with Ocean Productivity. Environ Microbiol 2010, 12, 490–500. [CrossRef]
- Tam, R.; Saier, M.H. Structural, Functional, and Evolutionary Relationships among Extracellular Solute-Binding Receptors of Bacteria. Microbiol Rev 1993, 57, 320–346. [CrossRef]
- Jaskulak, M.; Grobelak, A.; Vandenbulcke, F. Effects of Sewage Sludge Supplementation on Heavy Metal Accumulation and the Expression of ABC Transporters in Sinapis Alba L. during Assisted Phytoremediation of Contaminated Sites. Ecotoxicol Environ Saf 2020, 197, 110606. [CrossRef]
- Giovannoni, S.J.; Tripp, H.J.; Givan, S.; Podar, M.; Vergin, K.L.; Baptista, D.; Bibbs, L.; Eads, J.; Richardson, T.H.; Noordewier, M.; et al. Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Science 2005, 309, 1242–1245. [CrossRef]
- Williams, T.J.; Ertan, H.; Ting, L.; Cavicchioli, R. Carbon and Nitrogen Substrate Utilization in the Marine Bacterium Sphingopyxis Alaskensis Strain RB2256. ISME J 2009, 3, 1036–1052. [CrossRef]
- Orsi, W.D.; Smith, J.M.; Liu, S.; Liu, Z.; Sakamoto, C.M.; Wilken, S.; Poirier, C.; Richards, T.A.; Keeling, P.J.; Worden, A.Z.; et al. Diverse, Uncultivated Bacteria and Archaea Underlying the Cycling of Dissolved Protein in the Ocean. ISME J 2016, 10, 2158–2173. [CrossRef]
- Lofton, M.E.; Brentrup, J.A.; Beck, W.S.; Zwart, J.A.; Bhattacharya, R.; Brighenti, L.S.; Burnet, S.H.; McCullough, I.M.; Steele, B.G.; Carey, C.C.; et al. Using Near-Term Forecasts and Uncertainty Partitioning to Inform Prediction of Oligotrophic Lake Cyanobacterial Density. Ecol Appl 2022, 32, e2590. [CrossRef]
- Chen, K.; Allen, J.; Lu, J. Community Structures of Phytoplankton with Emphasis on Toxic Cyanobacteria in an Ohio Inland Lake during Bloom Season. J Water Resour Prot 2017, 9, 1–29. [CrossRef]
- Rinta-Kanto, J.M.; Konopko, E.A.; Debruyn, J.M.; Bourbonniere, R.A.; Boyer, G.L.; Wilhelm, S.W. Lake Erie Microcystis: Relationship between Microcystin Production, Dynamics of Genotypes and Environmental Parameters in a Large Lake. Harmful Algae 2009, 8, 665–673. [CrossRef]
- Guo, S.; Wang, Y.; Huang, J.; Dong, J.; Zhang, J. Decoupling and Decomposition Analysis of Land Natural Capital Utilization and Economic Growth: A Case Study in Ningxia Hui Autonomous Region, China. Int J Environ Res Public Health 2021, 18, 646. [CrossRef]
- Peng, L.; Zhao, K.; Chen, S.; Ren, Z.; Wei, H.; Wan, C. Whole Genome and Acid Stress Comparative Transcriptome Analysis of Lactiplantibacillus Plantarum ZDY2013. Arch Microbiol 2021, 203, 2795–2807. [CrossRef]
- Zhang, L.; Song, D.; Wu, Z. Transcriptome Analysis of Cyclocarya Paliurus Flavonoids Regulation of Differently Expressed Genes in Enterococcus Faecalis under Low PH Stress. Arch Microbiol 2021, 203, 2147–2155. [CrossRef]
- Petrov, K.; Arsov, A.; Petrova, P. Butanol Tolerance of Lactiplantibacillus Plantarum: A Transcriptome Study. Genes (Basel) 2021, 12, 181. [CrossRef]
- Cheng, C.; Han, X.; Xu, J.; Sun, J.; Li, K.; Han, Y.; Chen, M.; Song, H. YjbH Mediates the Oxidative Stress Response and Infection by Regulating SpxA1 and the Phosphoenolpyruvate-Carbohydrate Phosphotransferase System (PTS) in Listeria Monocytogenes. Gut Microbes 2021, 13, 1–19. [CrossRef]
- Vasylkivska, M.; Jureckova, K.; Branska, B.; Sedlar, K.; Kolek, J.; Provaznik, I.; Patakova, P. Transcriptional Analysis of Amino Acid, Metal Ion, Vitamin and Carbohydrate Uptake in Butanol-Producing Clostridium Beijerinckii NRRL B-598. PLoS One 2019, 14, e0224560. [CrossRef]
- Stautz, J.; Hellmich, Y.; Fuss, M.F.; Silberberg, J.M.; Devlin, J.R.; Stockbridge, R.B.; Hänelt, I. Molecular Mechanisms for Bacterial Potassium Homeostasis. J Mol Biol 2021, 433, 166968. [CrossRef]
- Stülke, J.; Krüger, L. Cyclic Di-AMP Signaling in Bacteria. Annu Rev Microbiol 2020, 74, 159–179. [CrossRef]
- Fang, H.; Qin, X.Y.; Zhang, K.D.; Nie, Y.; Wu, X.L. Role of the Group 2 Mrp Sodium/Proton Antiporter in Rapid Response to High Alkaline Shock in the Alkaline- and Salt-Tolerant Dietzia Sp. DQ12-45-1b. Appl Microbiol Biotechnol 2018, 102, 3765–3777. [CrossRef]
- Dürre, P. Physiology and Sporulation in Clostridium. Microbiol Spectr 2014, 2, TBS-0010-2012. [CrossRef]
- Eisenstadt, E. Potassium Content during Growth and Sporulation in Bacillus Subtilis. J Bacteriol 1972, 112, 264–267. [CrossRef]
- Vohradsky, J.; Schwarz, M.; Ramaniuk, O.; Ruiz-Larrabeiti, O.; Vaňková Hausnerová, V.; Šanderová, H.; Krásný, L. Kinetic Modeling and Meta-Analysis of the Bacillus Subtilis SigB Regulon during Spore Germination and Outgrowth. Microorganisms 2021, 9, 112. [CrossRef]
- Stetsenko, A.; Guskov, A. Cation Permeability in CorA Family of Proteins. Sci Rep 2020, 10, 840. [CrossRef]
- Payandeh, J.; Pfoh, R.; Pai, E.F. The Structure and Regulation of Magnesium Selective Ion Channels. Biochim Biophys Acta 2013, 1828, 2778–2792. [CrossRef]
- Pohland, A.C.; Schneider, D. Mg2+ Homeostasis and Transport in Cyanobacteria - at the Crossroads of Bacterial and Chloroplast Mg2+ Import. Biol Chem 2019, 400, 1289–1301. [CrossRef]
- Trachsel, E.; Redder, P.; Linder, P.; Armitano, J. Genetic Screens Reveal Novel Major and Minor Players in Magnesium Homeostasis of Staphylococcus Aureus. PLoS Genet 2019, 15, e1008336. [CrossRef]
- Saha, J.; Dey, S.; Pal, A. Whole Genome Sequencing and Comparative Genomic Analyses of Pseudomonas Aeruginosa Strain Isolated from Arable Soil Reveal Novel Insights into Heavy Metal Resistance and Codon Biology. Curr Genet 2022, 68, 481–503. [CrossRef]
- Cheng, D.; He, Q. PfsR Is a Key Regulator of Iron Homeostasis in Synechocystis PCC 6803. PLoS One 2014, 9, e101743. [CrossRef]
- Cheng, Y.; Zhang, T.; Cao, Y.; Wang, L.; Chen, W. New Insights into the Function of the Proteins IsiC and IsiD from Synechocystis Sp. PCC 6803 under Iron Limitation. Appl Microbiol Biotechnol 2021, 105, 4693–4707. [CrossRef]
- Seo, S.W.; Kim, D.; Latif, H.; O’Brien, E.J.; Szubin, R.; Palsson, B.O. Deciphering Fur Transcriptional Regulatory Network Highlights Its Complex Role beyond Iron Metabolism in Escherichia Coli. Nat Commun 2014, 5, 4910. [CrossRef]
- Plante, S.; Labbé, S. Spore Germination Requires Ferrichrome Biosynthesis and the Siderophore Transporter Str1 in Schizosaccharomyces Pombe. Genetics 2019, 211, 893–911. [CrossRef]
- Pivato, M.; Ballottari, M. Chlamydomonas Reinhardtii Cellular Compartments and Their Contribution to Intracellular Calcium Signalling. J Exp Bot 2021, 72, 5312–5335. [CrossRef]
- Cao, R.; Qin, P.; Li, W.; Shang, C.; Chai, Y.; Jin, D.; Chen, A. Hydrogen Sulfide and Calcium Effects on Cadmium Removal and Resistance in the White-Rot Fungus Phanerochaete Chrysosporium. Appl Microbiol Biotechnol 2021, 105, 6451–6462. [CrossRef]
- Wan, Y.; Wang, M.; Chan, E.W.C.; Chen, S. Membrane Transporters of the Major Facilitator Superfamily Are Essential for Long-Term Maintenance of Phenotypic Tolerance to Multiple Antibiotics in E. Coli. Microbiol Spectr 2021, 9, e0184621. [CrossRef]
- Wani, A.K.; Akhtar, N.; Sher, F.; Navarrete, A.A.; Américo-Pinheiro, J.H.P. Microbial Adaptation to Different Environmental Conditions: Molecular Perspective of Evolved Genetic and Cellular Systems. Arch Microbiol 2022, 204, 144. [CrossRef]
- Coloma-Rivero, R.F.; Flores-Concha, M.; Molina, R.E.; Soto-Shara, R.; Cartes, Á.; Oñate, Á.A. Brucella and Its Hidden Flagellar System. Microorganisms 2021, 10, 83. [CrossRef]
- Zhou, M.; Liu, Z.; Wang, J.; Zhao, Y.; Hu, B. Sphingomonas Relies on Chemotaxis to Degrade Polycyclic Aromatic Hydrocarbons and Maintain Dominance in Coking Sites. Microorganisms 2022, 10, 1109. [CrossRef]
- Solar Venero, E.C.; Ricardi, M.M.; Gomez-Lozano, M.; Molin, S.; Tribelli, P.M.; López, N.I. Oxidative Stress under Low Oxygen Conditions Triggers Hyperflagellation and Motility in the Antarctic Bacterium Pseudomonas Extremaustralis. Extremophiles 2019, 23, 587–597. [CrossRef]
- Sridhar, J.; Gayathri, M. Transcriptome Based Identification of Silver Stress Responsive SRNAs from Bacillus Cereus ATCC14579. Bioinformation 2019, 15, 474–479. [CrossRef]
- Amin, S.V.; Roberts, J.T.; Patterson, D.G.; Coley, A.B.; Allred, J.A.; Denner, J.M.; Johnson, J.P.; Mullen, G.E.; O’Neal, T.K.; Smith, J.T.; et al. Novel Small RNA (SRNA) Landscape of the Starvation-Stress Response Transcriptome of Salmonella Enterica Serovar Typhimurium. RNA Biol 2016, 13, 331–342. [CrossRef]
- Landt, S.G.; Lesley, J.A.; Britos, L.; Shapiro, L. CrfA, a Small Noncoding RNA Regulator of Adaptation to Carbon Starvation in Caulobacter Crescentus. J Bacteriol 2010, 192, 4763–4775. [CrossRef]
- Babitzke, P.; Lai, Y.-J.; Renda, A.J.; Romeo, T. Posttranscription Initiation Control of Gene Expression Mediated by Bacterial RNA-Binding Proteins. Annu Rev Microbiol 2019, 73, 43–67. [CrossRef]
- Chioccioli, S.; Del Duca, S.; Vassallo, A.; Castronovo, L.M.; Fani, R. Exploring the Role of the Histidine Biosynthetic HisF Gene in Cellular Metabolism and in the Evolution of (Ancestral) Genes: From LUCA to the Extant (Micro)Organisms. Microbiol Res 2020, 240, 126555. [CrossRef]
- Fani, R.; Brilli, M.; Fondi, M.; Lió, P. The Role of Gene Fusions in the Evolution of Metabolic Pathways: The Histidine Biosynthesis Case. BMC Evol Biol 2007, 7 Suppl 2, S4. [CrossRef]
- Martínez-Guitián, M.; Vázquez-Ucha, J.C.; Álvarez-Fraga, L.; Conde-Pérez, K.; Lasarte-Monterrubio, C.; Vallejo, J.A.; Bou, G.; Poza, M.; Beceiro, A. Involvement of HisF in the Persistence of Acinetobacter Baumannii During a Pneumonia Infection. Front Cell Infect Microbiol 2019, 9, 310. [CrossRef]
- Kuba, M.; Neha, N.; De Souza, D.P.; Dayalan, S.; Newson, J.P.M.; Tull, D.; McConville, M.J.; Sansom, F.M.; Newton, H.J. Coxiella Burnetii Utilizes Both Glutamate and Glucose during Infection with Glucose Uptake Mediated by Multiple Transporters. Biochem J 2019, 476, 2851–2867. [CrossRef]
- Byer, T.; Wang, J.; Zhang, M.G.; Vather, N.; Blachman, A.; Visser, B.; Liu, J.M. MtlR Negatively Regulates Mannitol Utilization by Vibrio Cholerae. Microbiology (Reading) 2017, 163, 1902–1911. [CrossRef]
- Zheng, Z.; Jiang, T.; Zou, L.; Ouyang, S.; Zhou, J.; Lin, X.; He, Q.; Wang, L.; Yu, B.; Xu, H.; et al. Simultaneous Consumption of Cellobiose and Xylose by Bacillus Coagulans to Circumvent Glucose Repression and Identification of Its Cellobiose-Assimilating Operons. Biotechnol Biofuels 2018, 11, 320. [CrossRef]
- Aboulwafa, M.; Zhang, Z.; Saier, M.H. Protein-Protein Interactions in the Cytoplasmic Membrane of Escherichia Coli: Influence of the Overexpression of Diverse Transporter-Encoding Genes on the Activities of PTS Sugar Uptake Systems. Microb Physiol 2020, 30, 36–49. [CrossRef]
- Chen, Q.; Li, F.; Zuo, X.; Chen, J.; Qin, P.; Wang, C.; Xu, J.; Yang, D.; Xing, B.; Liu, Y.; et al. Reversible Domain Closure Modulates GlnBP Ligand Binding Affinity. PLoS One 2022, 17, e0263102. [CrossRef]
- Zhang, M.G.; Liu, J.M. Transcription of Cis Antisense Small RNA MtlS in Vibrio Cholerae Is Regulated by Transcription of Its Target Gene, MtlA. J Bacteriol 2019, 201, e00178-19. [CrossRef]
- Poorinmohammad, N.; Hamedi, J.; Masoudi-Nejad, A. Genome-Scale Exploration of Transcriptional Regulation in the Nisin Z Producer Lactococcus Lactis Subsp. Lactis IO-1. Sci Rep 2020, 10, 3787. [CrossRef]
- Jang, H.; Kim, S.T.; Sang, M.K. Suppressive Effect of Bioactive Extracts of Bacillus Sp. H8-1 and Bacillus Sp. K203 on Tomato Wilt Caused by Clavibacter Michiganensis Subsp. Michiganensis. Microorganisms 2022, 10, 403. [CrossRef]
- Cao, T.N.; Joyet, P.; Aké, F.M.D.; Milohanic, E.; Deutscher, J. Studies of the Listeria Monocytogenes Cellobiose Transport Components and Their Impact on Virulence Gene Repression. J Mol Microbiol Biotechnol 2019, 29, 10–26. [CrossRef]
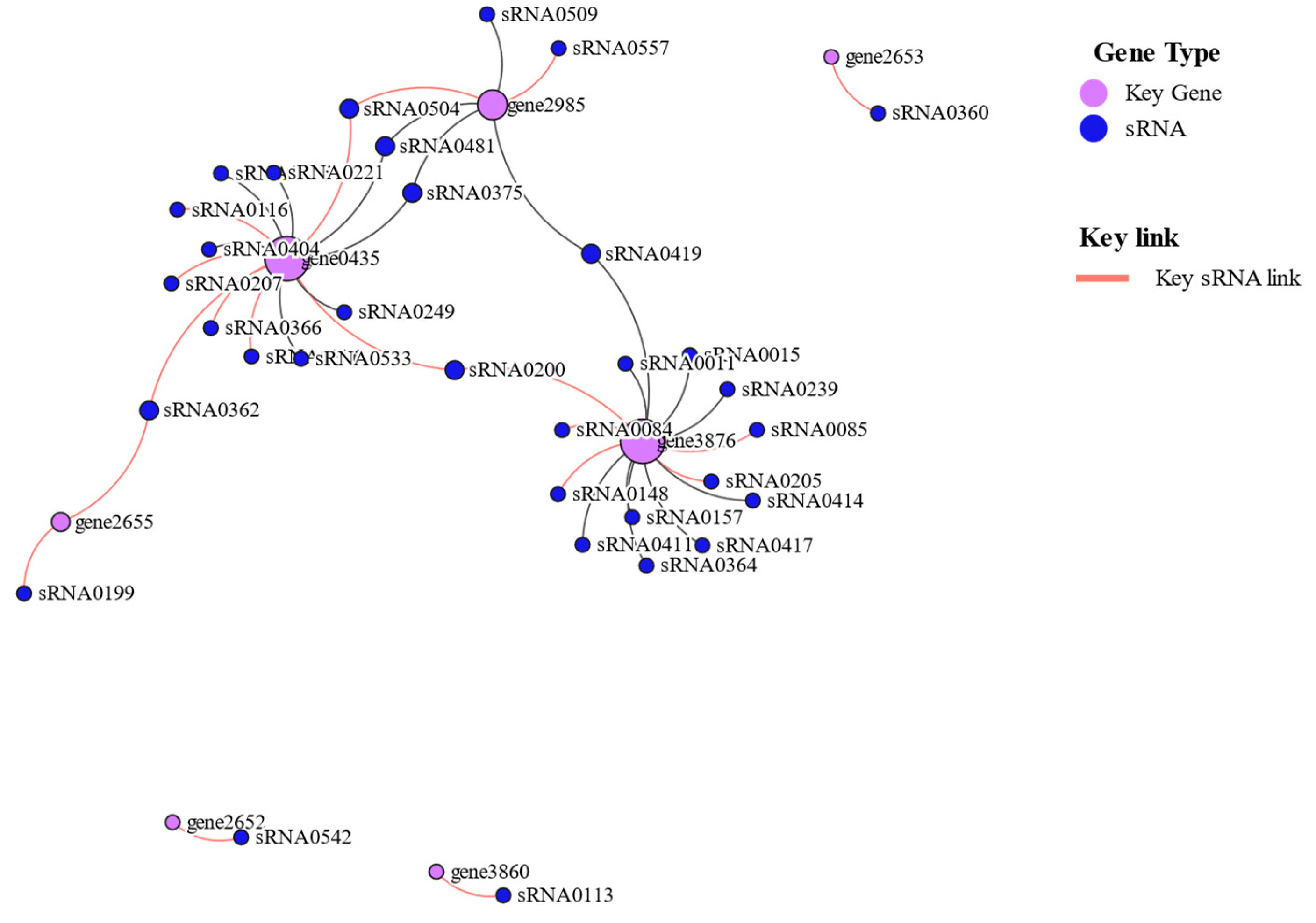
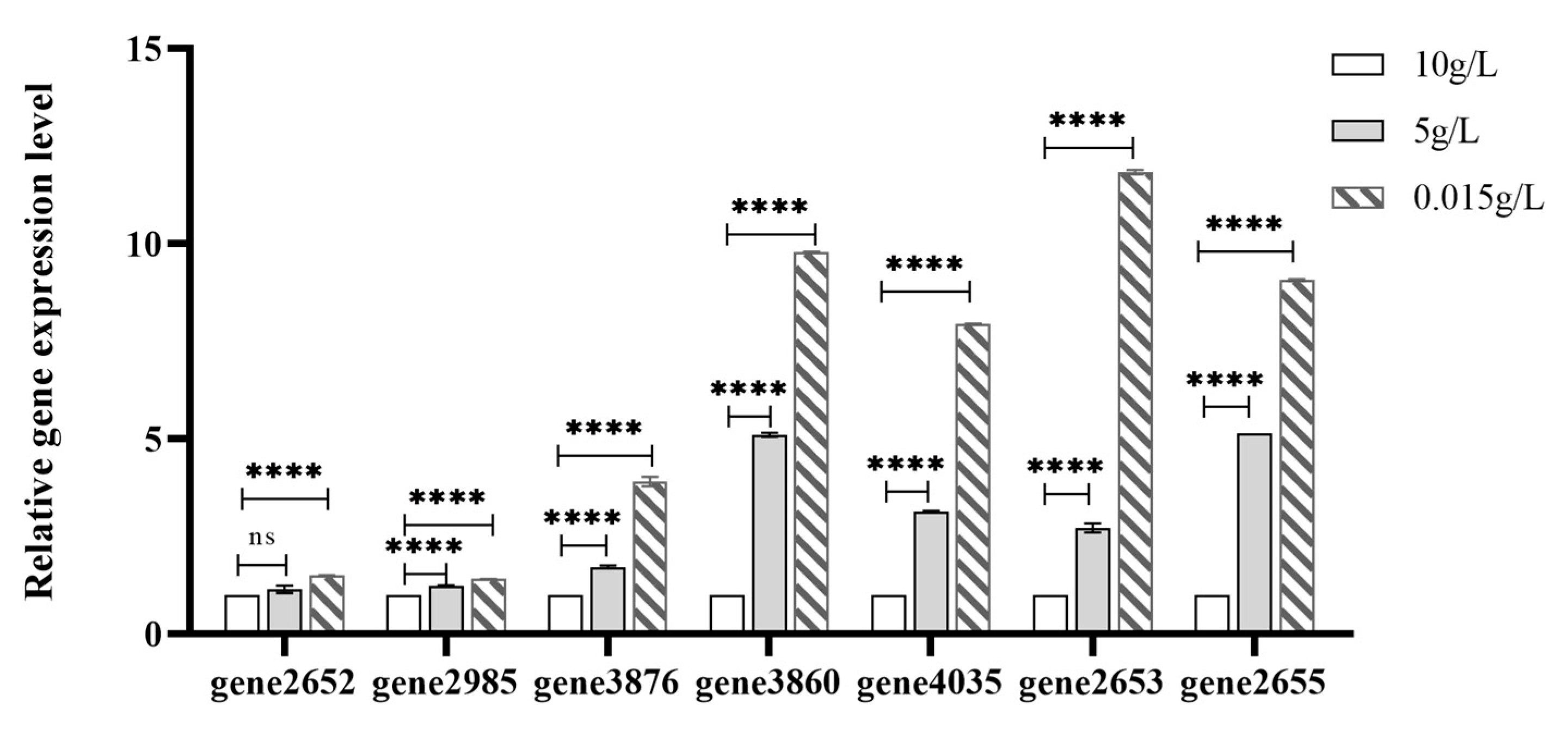
| Key Gene ID | Description | Gene Name | Log2FC(L/H) |
| gene2652 | amino acid ABC transporter ATPase | / | 7.448 |
| gene2985 | ABC transporter permease | / | 10.238 |
| gene3860 | oligo-beta-mannoside permease IIC protein | celB | 9.383 |
| gene0435 | PTS mannitol transporter subunit IIB | mtlA | 7.98 |
| gene3876 | oligo-beta-mannoside permease IIC protein | celB | 7.362 |
| gene2653 | glutamine ABC transporter substrate-binding protein | / | 7.496 |
| gene2655 | glutamine ABC transporter permease | / | 5.051 |
| sRNA_id | Log2FC(L/H) | Target Gene ID |
| sRNA0200 | 11.71890433 | gene0435;gene3876 |
| sRNA0113 | 11.533039 | gene3860 |
| sRNA0084 | 11.40169888 | gene3876 |
| sRNA0085 | 11.40169888 | gene3876 |
| sRNA0148 | 11.40169888 | gene3876 |
| sRNA0207 | 10.90597229 | gene0435 |
| sRNA0205 | 9.776851885 | gene3876 |
| sRNA0366 | 8.533373277 | gene0435 |
| sRNA0336 | 6.681651624 | gene0435 |
| sRNA0362 | 6.434679786 | gene0435;gene2655 |
| sRNA0116 | 6.373911204 | gene0435 |
| sRNA0360 | 6.357940855 | gene2653 |
| sRNA0542 | 6.212579528 | gene2652 |
| sRNA0557 | 6.026353075 | gene2985 |
| sRNA0199 | 5.159987109 | gene2655 |
| sRNA0504 | 5.048301125 | gene2985;gene0435 |
| sRNA0015 | 4.964673587 | gene3876 |
| sRNA0533 | 4.896095911 | gene0435 |
| sRNA0419 | 4.801798083 | gene2985;gene3876 |
| sRNA0239 | 4.618899947 | gene3876 |
| sRNA0249 | 4.578804747 | gene0435 |
| sRNA0495 | 3.975578933 | gene0435 |
| sRNA0411 | 3.854669068 | gene3876 |
| sRNA0509 | 3.764795398 | gene2985 |
| sRNA0221 | 3.520639327 | gene0435 |
| sRNA0414 | 3.022561069 | gene3876 |
| sRNA0481 | 2.967066947 | gene0435、gene2985 |
| sRNA0417 | 1.934699587 | gene3876 |
| sRNA0364 | 1.373518955 | gene3876 |
| sRNA0011 | 1.103524326 | gene3876 |
| sRNA0157 | -1.52566018 | gene3876 |
| sRNA0404 | -1.81557978 | gene0435 |
| sRNA0375 | -2.395421117 | gene2985;gene0435 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




