1. Introduction
Coronavirus (CoV) belongs to Orthocoronavirinae subfamily,
Coronaviridae family in the
Nidovirales order. CoV is an enveloped, positive-sensed, single-stranded RNA virus with a genome length around 30000 bp, which is the largest RNA genome of virus discovered so far [
1]. According to the International Committee on Taxonomy of Viruses (ICTV) classification criteria based on the five structural domains of CoV polyprotein 1ab (pp1ab): RNA dependent RNA polymerase (RdRp), Nidovirus RdRp-associated nucleotidyltransferase (NiRAN), 3-chymotrypsin-likepro-tease (3CL-pro), helicase of superfamily 1 (HEL1) and zinc-binding domain (ZBD), the subfamily Orthocoronavirinae was classified into four genera (52 species and 26 subgenera):
Alphacoronavirus (α-CoV) including 26 species and 15 subgenera,
Betacoronavirus (β-CoV) including 14 species and 5 subgenera,
Deltacoronavirus (δ-CoV) including 7 species and 3 subgenera, and
Gammacoronavirus (γ-CoV) including 5 species and 3 subgenera (
htpps://ictv.global.taxonomy). CoVs show host specificity and tissue infection preference. Typically, α-CoV and β-CoV infect mammals; γ-CoV and δ-CoV mainly infect birds, some of which can also infect mammals [
2]. Since Hamre [
3] et al. discovered human coronavirus 229E (HCoV-229E) in the United States in 1966, a total of seven human coronaviruses (HCoVs) have been identified, besides HCoV-229E, the remaining six human coronaviruses are: NL63 (HCoV-NL63), OC43 (HCoV-OC43), HKU1 (HCoV-HKU1), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Among above HCoVs, HCoV-229E, OC43, NL63, and HKU1 are more common in the population, less pathogenic, and normally cause only mild respiratory symptoms similar to the common cold. However, emerging HCoVs inclduing SARS-CoV in 2002–2003, MERS-CoV in 2012, SARS-CoV-2 in 2019 show high pathogenicity characteristics. Especially the pandemic SARS-CoV-2 caused a serious public health issue of global concern. According to known knowledge, all HCoVs have been found to have related prototypes in animals: HCoV-OC43 and HKU1 related strains in from rodents [
4], CoVs similar to SARS-CoV, MERS-CoV, HCoV-NL63, HCoV-229E, and SARS-CoV-2 in bats [
5,
6]; Except for SARS-CoV, MERS-CoV, and HCoV-OC43, whose intermediate hosts may be civet [
7,
8,
9], dromedaries [
10] and cattle [
11], respectively, it is not well understood how the remaining four CoVs transmit the virus to humans. The high mutation characteristics of CoV present a high degree of genetic diversity while promoting its transmission.
CoV is widespread in nature that plays an important position in emerging infectious diseases. Zoonotic CoV was discovered in the 1960s [
12], and the frequency and scope of influence are increasing. The vast majority of animal infectious diseases originate from wildlife, and rodents represented by rats carry a variety of zoonotic pathogens, which play an extremely important role in emerging zoonoses. In many places, rodents come into close contact with humans, farm animals or pets. Rodents in towns and around cities provide a nexus between wildlife communities and humans, exposing humans to some zoonoses circulating in these natural ecosystems [
13]. The mouse coronavirus has been associated with rodents, and the prototype virus, firstly named Murine hepatitis virus (MHV) renamed
Murine coronavirus by ICTV, was the first isolated in mice in 1949 [
14]. In 1970, a variant was discovered in rats called the rat salivary gland-nucleitis coronavirus [
15]. It was not until 2014 that a new Murine coronavirus,
Lucheng Rn rat coronavirus (LRNV) from Norway Rat (
Rattus norvegicus) was discovered, as well as two new variants of β-CoV-
Longquan Aa mouse coronavirus (LAMV) from striped field mouse (
Apodoses agrarius) and
Longquan Rl rat coronavirus (LRLV) from lesser rice field rat (
Rattus. Lossea) [
16]. The CoVs found in rodents are divided into two lineages: the A lineage of β-CoV and a separate lineage of α-CoV [
17]. Then in 2015, a new Murine coronavirus,
China Rattus HKU24, belonging to the A lineage of β-CoV was discovered in Norway Rat in China [
4]. In 2022, a novel coronavirus,
Myodes coronavirus 2JL14 was reported from Swedish Bank Voles (
Myodes glareolus) [
18]. Recently, two new CoVs of
Suncus murinus coronavirus X74 from Asian house shrews and
Sorex araneus coronavirus T14 from common shrew were discovered [
19,
20]. In Yunnan Province of China, we have detected genetically diverse α-CoV and β-CoV in a variety of rats, such as Chevrieri’s field mouse (
Apodemus chevrieri) and large Chinese vole (
Eothenomys miletus), and identified the CoV genomes [
21]. All these findings suggest that small mammals, especially rodents may carry a wide range of CoVs.
In this study, we collected a total of 502 small mammals belonging to 18 species in 12 genera and 4 orders in Yunnan Province of China, and investigated the genetic diversity and epidemic of CoV by RT-PCR screening and sequencing of partial RdRp gene of CoV. The qRT-PCR method was further established for quantification of CoV, and the viral copy number of CoV was absolutely quantified, as well as tissue tropism of CoV in heart, liver, spleen, lung, kidney and intestinal tissues of RT-PCR positive samples. The results of this study increase our understanding of CoV diversity and provide a method for rapid, accurate and reliable screening of CoV.
2. Materials and Methods
2.1. Ethics Statement
The collection of small animals was performed by veterinarians with approval from the Animal Ethics Committee of Dali University (DLDXLL2020007).
2.2. Sample Collection and Processing
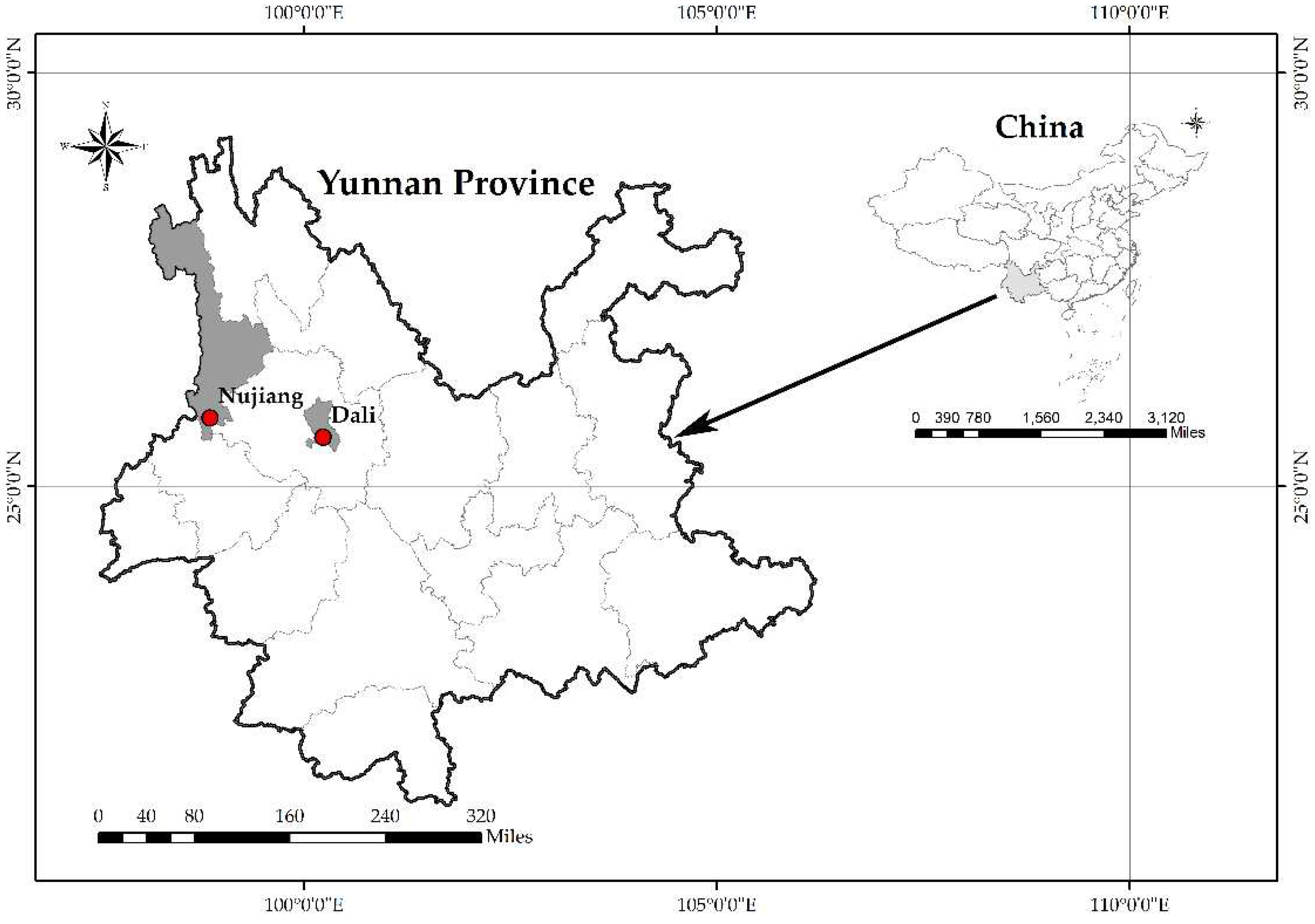
Small mammal samples were collected from August 2020 to August 2022 in Dali and Nujiang Prefecture, Yunnan Province, China (
Figure 1), using freshly fried fritters as bait. The samples trapping was performed using cage-type traps to capture, and the captured samples were collected at the destination early the next morning. The collected small mammals were brought back to the laboratory and euthanized, with species identification initially based on morphology, followed by further molecular identification of the species by sequence analysis of the mitochondrial (mt)-cytochrome b (
Cytb) gene [
22]. The samples were dissected in a sterile environment, and the heart, liver, spleen, lung, kidney, and intestinal tissues were collected in 2 mL cryogenic vials (CORNING, China) and stored temporarily in liquid nitrogen. The samples were then stored at −80 °C before further laboratory analyses.
2.3. DNA and RNA Extraction
Under aseptic conditions, approximately 1 g of intestine and other tissue samples were cut into GeneReady Animal PIII crushing tubes (Life Real, China), and 600 μL of sterilized phosphate Buffered Saline (PBS) was added, followed by grinding in a GeneReady Ultimate grinder (Life Real, China). The 300 μL supernatant of the ground tissue sample was added to the nucleic acid extraction or purification kit (MagaBio plus Virus DNA/RNA Purification Kit III, China), and the sample DNA/RNA was extracted in a fully automated nucleic acid extraction and purification instrument (BIOER, China) according to the instructions, dispensed and stored at −80 °C until further analysis.
2.4. Primary Screening of CoV and Amplification of Partial RdRp Fragments
Using semi-nested PCR (RT-PCR), amplification against conserved regions of RdRp gene of CoV (
Table 1) [
23]. The primers were synthesized by Shanghai Sangon Biotech, and the system was 25 μL for both rounds. The first round of RT-PCR was performed using the Fastking One Step RT-PCR kit (TIANGEN, China). The reaction system of the first round was: 2× Fasting One Step RT-PCR MasterMix 12.5 μL, 25× RT-PCR Enzyme Mix 1 μL, CoV-FWD3 and CoV-FWD4/other (20 μM) 1 μL each, RNase-Free ddH
2O 6.5 μL, RNA template was 3 μL. First round PCR conditions were as follows: 42 °C, 30 min reverse transcription, followed by 35 cycles at 95 °C, 3 min pre-denaturation, 94 °C, 30 s denaturation, 48 °C, 30 s 35 annealing, 72 °C, 30 s elongation and a final extension 72 °C, 5 min, 10 °C, 1 min cooling. The second round PCR was performed with 2× Phanta Max Master Mix (Vazyme, China), and the reaction system was: 2× Phanta Max Master Mix 12.5 μL, 1 μL each of CoV-RVS3 and CoV-FWD4/other (20 μM), 9.5 μL of RNase-Free ddH
2O, and 1 μL of the product of the first round PCR was the template. Second round PCR conditions were as follows: 95 °C, 3 min pre-denaturation, 35 cycles at 94 °C, 15 s denaturation, 50 °C, 15 s annealing, 72 °C, 30 s extension and a final extension of 72 °C, 5 min, 10 °C, 1 min cooling. The length of the amplification product after two rounds of RT-PCR is approximately 443 bp. The second round of RT-PCR products were identified by agarose gel electrophoresis, and the positive amplification products that matched the expected size were purified by gel cutting (OMEGA Bio-tek, USA) and sent to Sangon Biotech for bi-directional sequence determination. For well-sequenced positive samples, specific primers were designed to amplify part of the ORF1ab fragment by multiple sequence alignment with the published coronavirus genome. To exclude PCR contamination, positive samples were verified by two independent PCRs performed by two different experimenters.
2.5. Virus Sequence Identification and Phylogenetic Analysis
Sequences were assembled by the DNAstar Lasergene software package and manually edited and cut to generate the final sequence of the viral gene. Similarity matching analysis was performed using the National Center for Biotechnology Information (NCBI) online Basic Local Alignment Search Tool (BLAST) based search tool. The CoV reference sequence set representing the RdRp gene was downloaded from GenBank, and sequence comparison was performed using ClustalX2. All viral sequences were constructed using the maximum likelihood method in MEGAX64 with a total of 1000 bootstrap replicates to generate. The evolutionary distances were computed using the Kimura 2-parameter method, and a self-spread value greater than 70% was generally considered a reliable evolutionary branching, and visualized in iTOL. The same method was used to construct the evolutionary tree based on the mt-Cytb gene for the corresponding small mammalian hosts. The α-CoV and β-CoV sequences in this study have been deposited to GenBank under the following numbers: OR223161-OR223180. The mt-Cytb gene sequences from small mammals in this study have been deposited to GenBank under the following numbers: OR223181-OR223200.
2.6. Construction of Plasmids and Determination of Virus Copies
The partial ORF1ab fragments of α-CoV and β-CoV representative strains were cloned into the pEASY-T1 vector (TRAN, China), and the T-loaded products were transformed into DH5α
E. coli cells. The inserted target genes were confirmed by sequencing after bacteriophage amplification. Small amounts of plasmids were extracted using the Plasmid Mini Kit I (Omega Bio-tek, USA) and stored at −80 °C in separate devices. Then, the concentration of the extracted coronavirus plasmid was determined by ultraviolet spectrophotometer (Life Real, China) after gradual thawing, and then according to the Equation (1), the plasmid concentration was converted to copies for the establishment of standard curves and quantitative analysis as a positive control.
2.7. Primer and TaqMan Probe Design and Optimization
After aligning part of the amplified ORF1ab fragment with the existing reference sequence in GeneBank, different specific primers and TaqMan probes were designed for the conserved sequences within the ORF1ab gene of α-CoV and β-CoV, respectively (
Table 1). Positive standards were diluted with RNase-Free ddH
2O in a 10-fold gradient and then used as templates for condition optimization and stored at −20 °C. The primer and TaqMan probe concentrations and annealing temperature within the qRTPCR system were optimized by several experiments to determine the optimal amplification conditions and reaction system.
HiScript® II U+ One Step qRT-PCR Probe Kits (Vazyme, China) were used. After several experiments to optimize the conditions, the reaction system for qRT-PCR was: 2× One Step U+ Mix 10 μL, One Step U+ Enzyme Mix 1 μL,50× ROX Reference Dye 2 0.4 μL, primers for both α-CoV and β-CoV 0.4 μL (10 μM), TaqMan probe 0.2 μL (10 μM), RNase-Free ddH2O 5.6 μL, and RNA template 2 μL. The reaction system for the assay was 20 μL, and the amplification reaction was performed using the Applied Biosystems 7500 Real-Time PCR system (Thermo Fisher Scientific, USA). The amplification conditions were as follows: 55 °C, 15 min reverse transcription, 45 cycles at 95 °C, 30 s pre-denaturation, 95 °C 10 s denaturation, α-CoV and β-CoV annealing temperatures and fluorescence signal acquisition times of 51 °C, 34 s and 60 °C, 34 s, respectively.
2.8. Establishment of Standard Curves
The positive standards diluted in a 10-fold gradient were used as templates and each concentration was repeated three times, the average of the three times was taken. The standard curve was plotted using the logarithm of the copies of the positive standard as the horizontal coordinate and the cycle threshold (Ct) value corresponding to the assay as the vertical coordinate, and the slope and correlation coefficient were calculated.
2.9. Evaluation of qRT-PCR Methods
2.9.1. Sensitivity
The limit of detection for qRT-PCR is determined by detecting a positive standard of serial dilution. α-CoV and β-CoV plasmid DNA concentrations were 356.828 ng/µL, 312.525 ng/µL, respectively, and the concentrations before dilution of the two genera were 6.09 × 1010 copies/µL, 5.24 × 1010 copies/µL, respectively. The plasmids were serially diluted 10-fold using RNase-Free ddH2O and used as qRT-PCR templates.
2.9.2. Specificity
To assess the specificity of qRT-PCR, Hantavirus, Orientia tsutsugamushi and Hepatitis E virus provided by our laboratory were compared with CoV.
2.9.3. Repeatability and Stability
Six concentration gradients (1.00 × 104–1.00 × 109 copies/µL) of positive standards diluted in a 10-fold gradient were used as templates, and each concentration was repeated 3 times as intra-group replicates, and the above operation was performed once a week for a total of 3 times as inter-group replicates. RNase-Free ddH2O was used as a negative control group for testing the intra-group and inter-group variation of different concentrations of positive standards. The mean Ct value (Mean Ct), standard deviation (SD) and coefficient of variation (CV) were calculated to evaluate the reproducibility and stability of qRT-PCR.
2.9.4. Comparison of qRT-PCR and RT-PCR Sensitivity
RT-PCR reactions were performed using serial 10-fold gradient dilutions of positive standards (1.00 × 109–1.00 × 101 copies/µL) as positive templates and RNase-free H2O as negative controls. The amplified PCR products were subjected to agarose gel electrophoresis to compare the sensitivity of qRT-PCR and RT-PCR methods.
2.10. qRT-PCR for Small Mammalian Samples
After CoV RNA extraction from tissues of small mammals captured in Dali and Nujiang Prefecture of Yunnan from August 2020 to August 2022 according to the above method, all samples were tested for α-CoV and β-CoV respectively according to the optimized qRT-PCR experimental conditions.
2.11. Tissue Tropism of Small Mammalian CoV
The samples tested positive by qRT-PCR were extracted from the heart, liver, spleen, lung and kidney tissues according to the requirements of the nucleic acid extraction kit and instruments, and then the tissues of small mammalian samples with α-CoV and β-CoV positives were quantitatively analyzed to study their tissue tropism.
2.12. Statistical Analysis
Data were analyzed using a one-way ANOVA with GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA). All results are expressed as the mean ± standard error of the mean (SEM). P-values < 0.05 were considered statistically significant, while P-values < 0.001 (three-star sign) and 0.0001 (four-star sign) were considered highly significant.
3. Results
3.1. Collection of Samples and Detection of CoV
A total of 502 small mammals of 18 species in 12 genera and 4 orders were collected in residential areas, arable areas and wild bush areas in Dali and Nujiang Prefecture of Yunnan province (
Table 2). RT-PCR was used to detect α-CoV and β-CoV RNA based on partial RdRp sequences. The overall positives of CoV was 20 including β-CoV (13) and α-CoV (7) with 3.98% prevalence in intestinal tissue samples. The prevalence of β-CoV was 3.54% (4/113) and 6.67% (6/90) in Chevrieri’s field mouse (
Apodemus chevrieri) and Lancangjiang field mouse (
A. ilex), respectively. The prevalence of both β-CoV and α-CoV in Kachin red-backed vole (
Eothenomys cachinus) and Norway rat (
Rattus norvegicus) was 2.22% (2/90), 3.85% (1/26), respectively. The prevalence of α-CoV in White-footed Indochinese rat (
R. nitidus) and long-tailed red-toothed shrew (
Episoriculus leucops) was 75% (3/4), and 5.88% (1/17), respectively (
Table 2).
3.2. Comparison of Partial RdRp gene
The amplified partial RdRp sequences of the 13 strains of β-CoV in this study shared 83.42%–99.23% nucleotide (nt) identity and 90.14%–100.00% amino acid (aa) identity, respectively (
Table S1). Among them, CoVDL55 (OR2233161), CoVDL75 (OR223162) from
A. chevrieri and CoVNJ21, CoVNJ33, CoVNJ53 (OR223169, OR223170, OR223172) from
A. ilex had the highest identity with Lijiang-41 (MT820628) from Sichuan field mouse (
Apodemus latronum) in Lijiang, China, and the nt identity was 96.43%–99.15%; CoVDL161 (OR223165), CoVDL172 (OR223166) from
A. chevrieri had the highest identity with Lijiang-53 (MT820629) from
A. chevrieri in Lijiang, China, with the nt identity of 97.19%–98.28%; CoVDL140 (OR223164) from
R. norvegicus and CoVNJ55 (OR223173) from
A. ilex had the highest identity with Ruili-874 (MT820631) from
R. tanezumi in Ruili, China, with the nt identity of 99.23%–99.57%; CoVNJ16 (OR223168), CoVNJ56 (OR223174) from
A. ilex had the highest identity with RtAp/SAX2015 (KY370064) from
A. peninsulae in Shanxi, China, with the nt identity of 96.29–98.07%; CoVNJ99 (OR223175), CoVNJ142 (OR223176) from
E. cachinus had the highest identity with BOV-36/IND/2015 (MH753496) from Indian bovine and DcCoV-HKU23 (MN514976) from one-humped camel (
Camelus dromedaries) in Morocco, with the nt identity of 97.86%–98.33%. The identity of nt, aa levels of the 7 α-CoV positive sequences was 94.00%–99.18% and 94.44%–99.31%, respectively (
Table S2). CoVDL82 (OR223163) from
R. norvegicus, CoVNJ135 (OR223176) from
Ep. Leucops, CoVNJ195, CoVNJ196 and CoVNJ207 (OR223178-OR223180) from
R. nitidus had the highest identity with RtRl/FJ2015 (KY370050) from
R. losea in Fujian, China, with the nt identity of 98.08%–98.97%; CoVNJ3 (OR223167), CoVNJ52 (OR223171) from
E. cachinus had the highest identity with RtClan/GZ2015 (KY370054) from
Eothenomys melanogaster, in Guizhou, China, with the nt identity of 93.23%–95.48%.
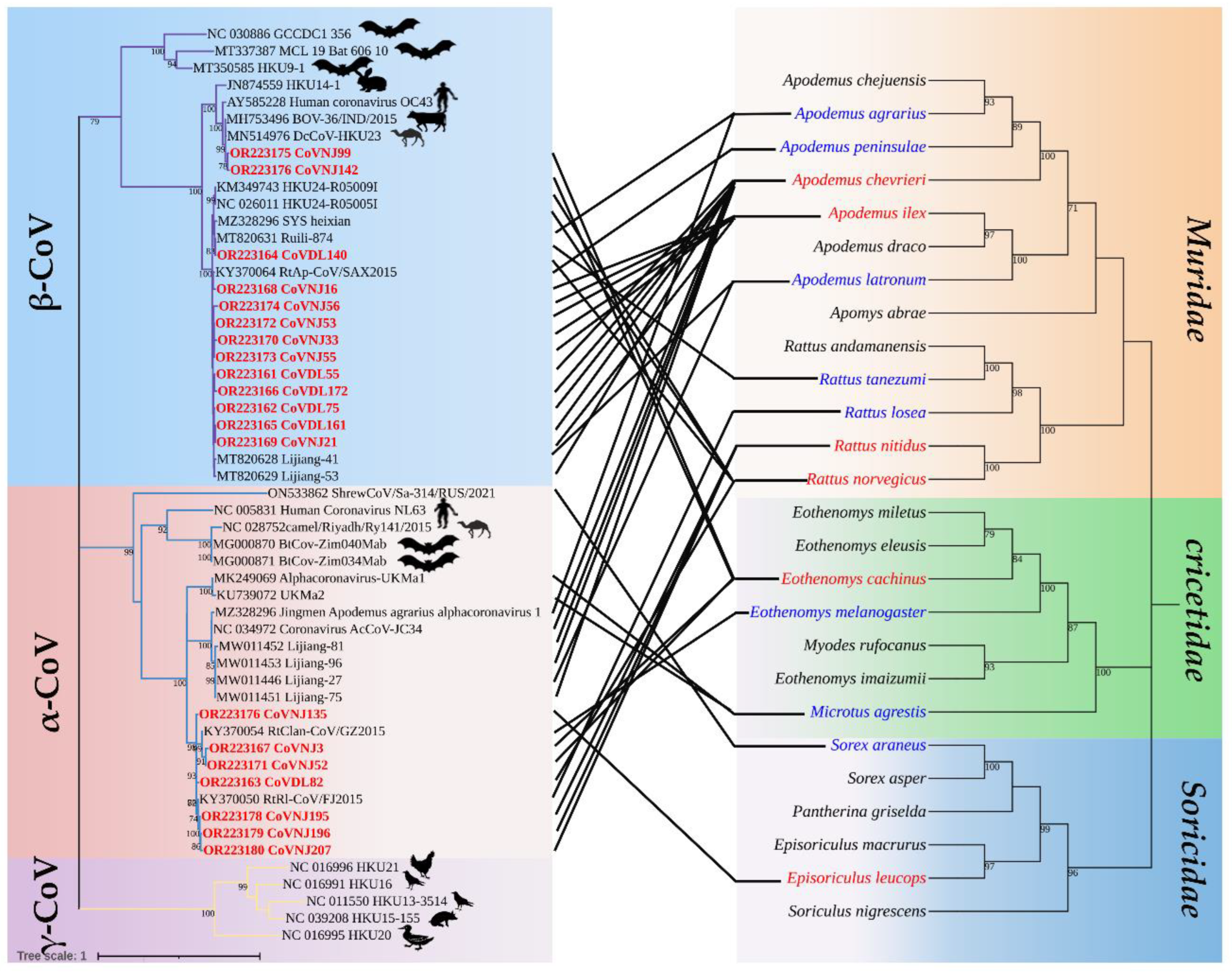
3.3. Phylogenetic Analysis
Among the 20 strains of CoVs identified in this study, CoVDL140 from
R. norvegicus (OR223184), CoVDL55, CoVDL75, CoVDL161, CoVDL172 from
A. chevrieri (OR223181, OR223182, OR223185, OR223186) and CoVNJ21, CoVNJ33, CoVNJ53, CoVNJ55 from
A. ilex (OR223189, OR223190, OR223192, OR223193) were clustered with the
China Rattus HKU24 representative strains Ruili-874, Lijiang-41, Lijiang-53, respectively, belonging to the
Embecovirus subgenus of β-CoV. CoVNJ16 and CoVNJ56 from the
A. ilex (OR223188, OR223194) was more closely related to RtAp/SAX2015; CoVNJ99 and CoVNJ142 from the
E. cachinus (OR223195, OR223197) clustered together with BOV-36/IND/2015 from Indian Bovine and DcCoV-HKU23 from
Camelus dromedarius in Morocco. Through the phylogenetic tree, it showed that HCoV-OC43 is also closely related to CoVNJ99 and CoVNJ142. CoVNJ3 and CoVNJ52 from
E. cachinus (OR223187, OR223191), CoVDL82 from
R. norvegicus (OR223183), CoVNJ135 from
Ep. Leucops (OR223196), CoVNJ195, CoVNJ196 and CoVNJ207 from
R. nitidus (OR223198-OR223200) are more closely related to RtRl/FJ2015 and RtClan/GZ2015, all of which are α-CoV, and viruses with high similarity were found in insectivores and rodents. In addition, identical viruses can be identified in multiple species from the same geographical location, for example, CoVNJ135 from
Ep. leucops and CoVNJ195, CoVNJ196, and CoVNJ207 from
R. nitidus in Nujiang Prefecture are the most similar to RtRl/FJ2015 (
Figure 2).
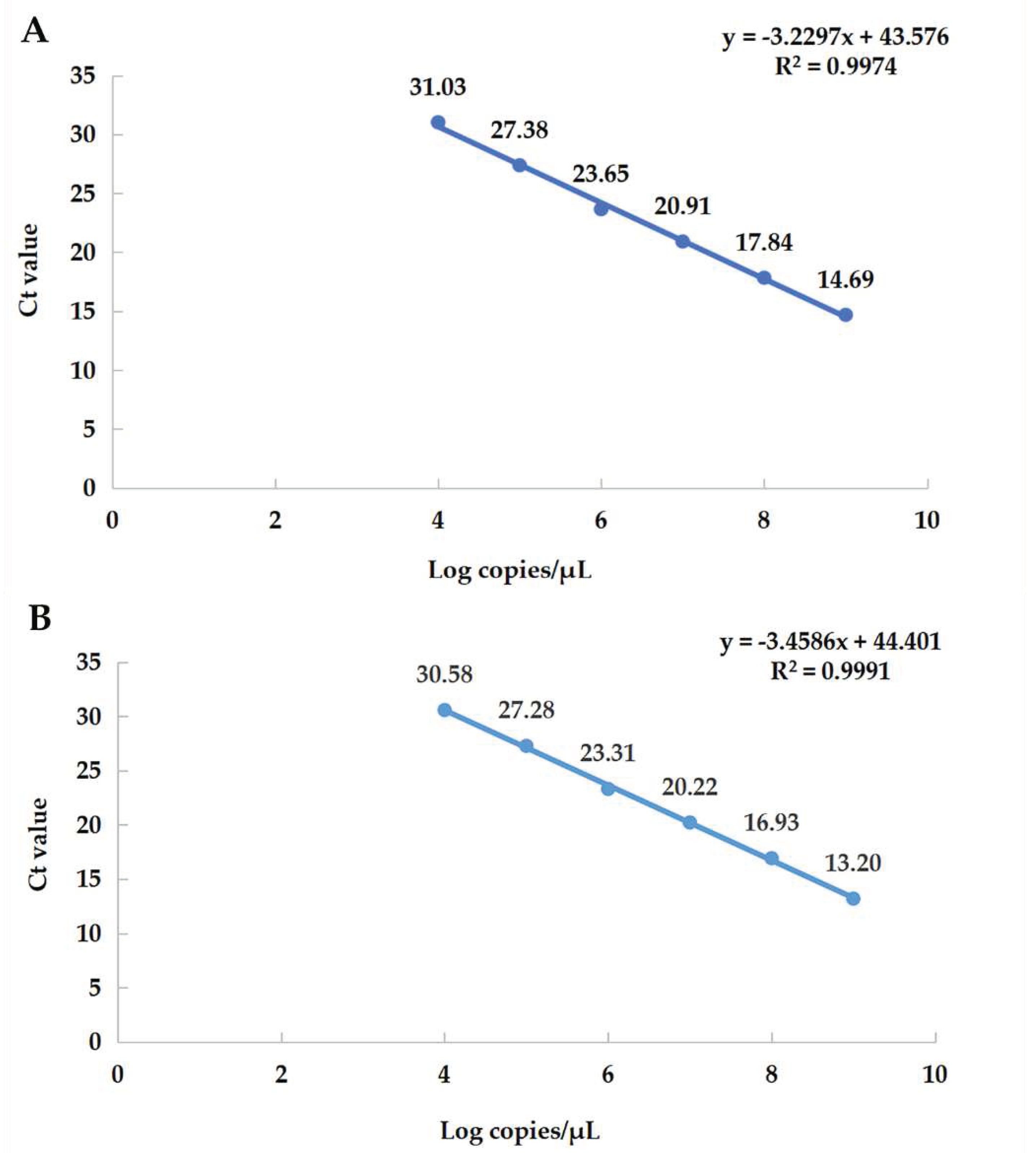
3.4. Establishment of qRT-PCR Standard Curves
The six consecutive dilution gradients of α-CoV and β-CoV standards (1.00 × 10
4–1.00 × 10
9 copies/μL) were selected as the log values of copy number on the X axis, and the obtained Ct values were plotted as the standard curve on the Y axis (
Figure 3). It shows that the template of the gradient dilution has a good linear relationship with the Ct value.
3.5. Evaluation of qRT-PCR Methods
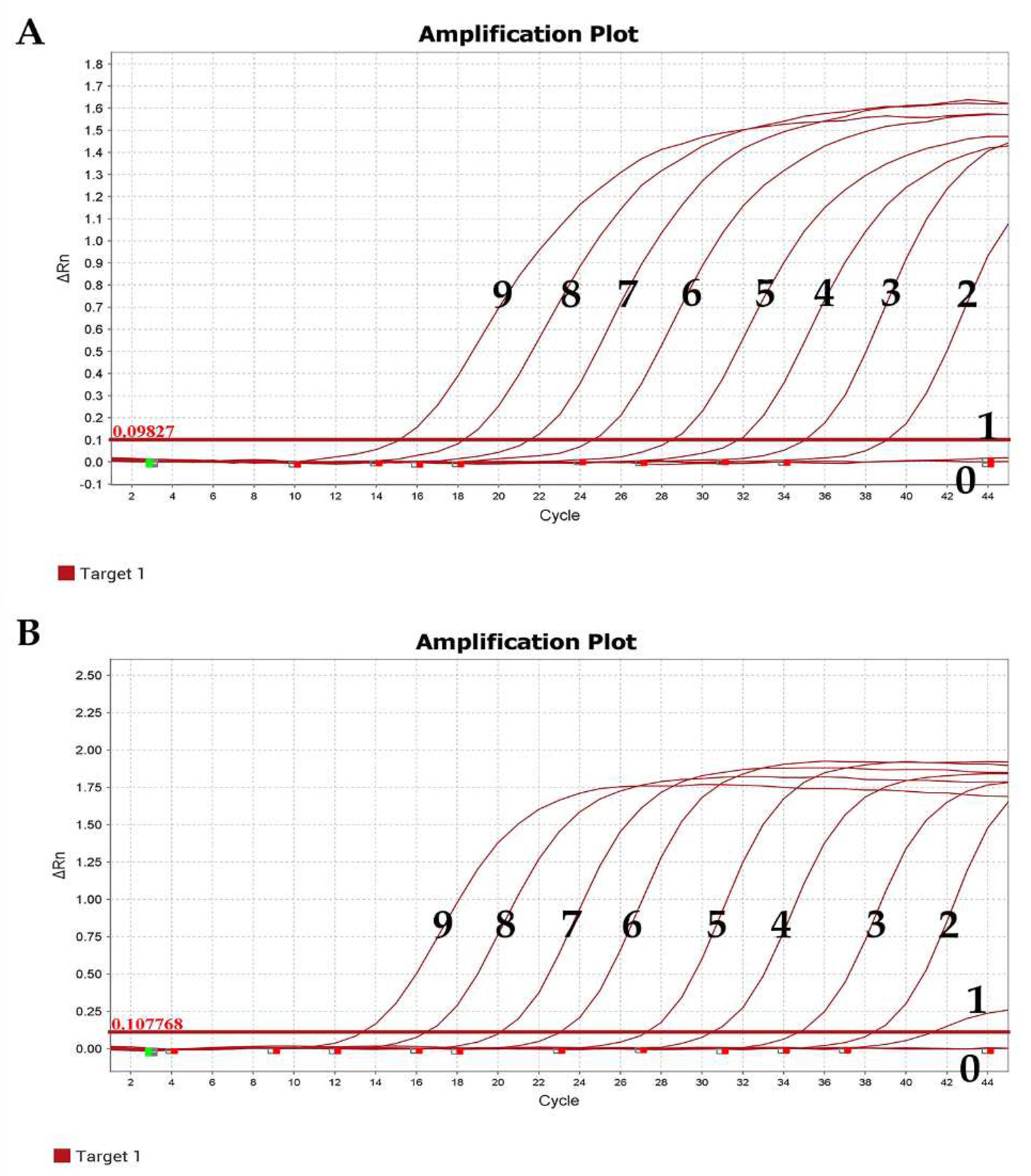
3.5.1. Sensitivity
The experimental results showed that the minimum copy number detectable by positive standards for both α-CoV and β-CoV was 1.00 × 10
1 copies/µL, indicating the good sensitivity of the established qRT-PCR method (
Figure 4).
3.5.2. Specificity
The results of the experiments showed that the specific primers designed only showed amplification curves for the positive standards of α-CoV and β-CoV, and no amplification curves and no fluorescence signal in the negative control group, suggesting that the established qRT-PCR method had good specificity.
3.5.3. Repeatability and Stability
Intra-group repeatability test: The experimental results showed that the SD in the standard group for each concentration of α-CoV was between 0.06–0.30 and CV was between 0.26–1.09. The SD in the standard group for each concentration of β-CoV was between 0.05–0.39 and CV between 0.16–1.66 (
Table S3).
Inter-group repeatability test: The experimental results showed that the SD in the standard group for each concentration of α-CoV was between 0.01–0.16, and CV was between 0.03–0.96. The SD in the standard group for each concentration of β-CoV was between 0.04–0.47, and the CV was between 0.22–1.77 (
Table S4). All these data indicated that the established qRT- PCR method has good reproducibility and stability.
3.6. Comparison of qRT-PCR and RT-PCR Sensitivity
The minimum copy number detected by qRT-PCR for both α-CoV and β-CoV positive standards was 1.00 × 10
1 copies/µL, while the minimum copy number detected by RT-PCR for α-CoV and β-CoV positive standards was 1.00 × 10
3 copies/µL, 1.00 × 10
4 copies/µL, respectively, which were 10
2 and 10
3 times higher than those of RT-PCR, respectively (
Figure 5).
3.7. Tissue Tropism
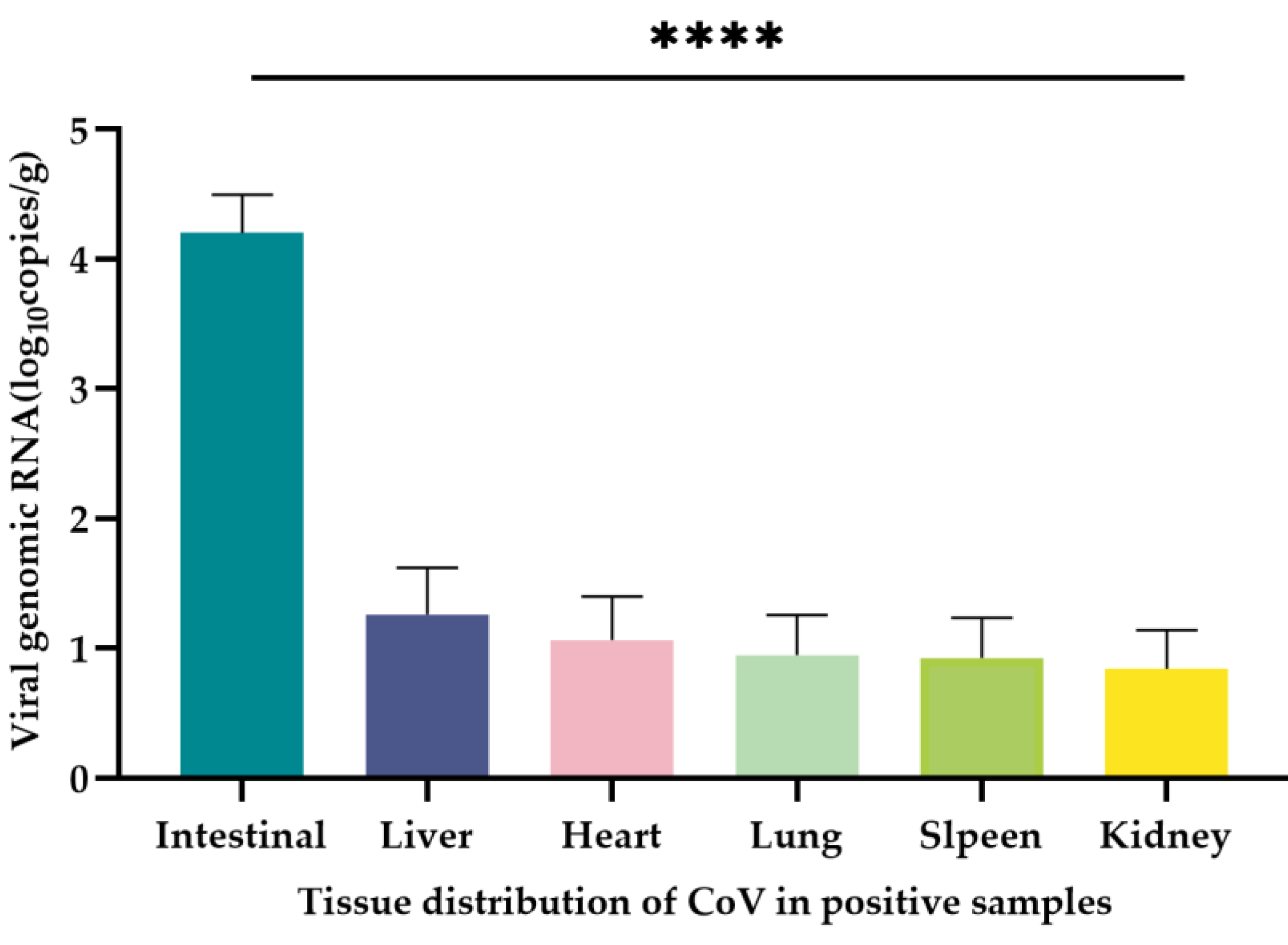
A total of 20 CoV-positive samples from Dali and Nujiang Prefecture of Yunnan Province, and CoV RNA naturally infected in the heart, liver, spleen, lung, kidney and intestinal tissues of the positive samples were quantified using the qRT-PCR method established in this study. The mean CoV copy number was 1.25 × 10
6 copies/g in the intestines of all positive samples with the highest CoV copy numbers. The mean CoV copy numbers in liver, heart, spleen, lung and kidney tissues were 4.02 × 10
3 copies/g, 2.89 × 10
3 copies/g, 2.88 × 10
3 copies/g, 1.68 × 10
3, and 1.29 × 10
3 copies/g, respectively (
Table 3). The intestinal tissue contained significantly higher viral copy numbers than that in liver, heart, spleen, lung and kidney tissues (P < 0.0001) (
Figure 6). The remaining tissues except the intestinal tissue also contained unequal copies of the virus, but there was no significant difference among tissues (P > 0.05).
4. Discussion
In this study, CoVs were detected in the six species of A. chevrieri, A. ilex, E. cachinus, R. norvegicus, R. nitidus and Ep. leucops. Among these hosts, β-CoV was found from three species of A. chevrieri, A. ilex and R. norvegicus, α-CoV was found from two species of R. nitidus and Ep. leucops, α-CoV and β-CoV co-infection was found in two species of E. cachinus and R. norvegicus. These results showed that the CoV is widely present and highly diverse in hosts of Rodentia and Insectivora. Other genetic characteristics of the α-CoV and β-CoV detected here need further genomic sequencing analysis. A. chevrieri and A. ilex were infected with highly similar CoV, while A. chevrieri and A. ilex were from Dali and Nujiang Prefecture in Yunnan Province, respectively. Nujiang Prefecture is located on the western border of Yunnan Province, China, adjacent to Myanmar and connected to Dali Prefecture in the Southeast that suggested a co-evolutionary relationship among CoVs and host animals. From the CoV and host co-evolutionary tree, it was found that CoV of different genera could infect the same rodents and CoV of the same genera could infect different rodents, suggesting the existence of cross-species transmission of CoV carried by rodents inhabiting the same habitat.
CoVs can infect a wide range of host animals [
24], and the cross-species transmission of CoVs has caused multiple epidemics of infection and disease in animals and humans, which have seriously affected human productive life and public health. Notably, this study detected CoV associated with CoVs carried by
Camelus dromedarius and Bovine in
E. cachinus, which are intermediate hosts for the Middle East respiratory syndrome coronavirus. MERS-CoV is repeatedly detected in one-humpted camelus, and MERS-CoV isolated from one-humpted camelus is genetically and phenotypically similar to the CoV that infects humans, including the virus’s spike protein, suggesting that CoV in one-humpted camelus may be transmitted to human [
25]. True primary zoonotic infection is difficult to identify, and it is possible that the host or other intermediate host will be altered during transmission of infection to humans [
26]. However, to date, no investigations have been conducted on the presence of MERS-CoV or similar viruses in wild rodents [
27]. Bovines are often grazed by herders in Nujiang Prefecture, so it is speculated that rats may have been exposed to bovine feces and there is cross-species transmission, so that highly related CoV from bovine infections can be detected on rats. The emergence and evolution of CoVs in new hosts is caused by a variety of factors, such as recombination, horizontal transfer of genes, gene duplication, and shift of open reading frames, all of which accelerate their infection with new hosts and enhance their ability to adapt to new hosts [
28]. Although only partial fragments were obtained in this experiment, this CoV was detected in rats in Nujiang Prefecture, Yunnan Province, China, and the related research should be strengthened.
The qRT-PCR established in this study had good sensitivity, specificity, stability and reproducibility, and the highest CoV copy number (P < 0.0001) was found to be contained in small mammalian intestinal tissue by quantitative studies, revealing that CoVs infecting small mammals have intestinal tropics. However, unequal amounts of CoV copies were also detected in liver, heart, spleen, lung and kidney tissues, with mean values ranging from 1.29 × 10
3 to 4.02 × 10
3 copies/g, suggesting that CoV has a wide range of tissue tropism and may be transmitted by oral-fecal, urinary and respiratory routes, which also provides evidence for the transmission route of CoV. It also re-confirms that CoVs are respiratory, intestinal, hepatic and renal pathogens in animals and humans, and provides evidence for the clinical signs and symptoms of infected patients including respiratory, intestinal, hepatic and renal manifestations, as well as other forms of disease [
29]. In previous decades of research, different tissue orientations of rodent CoVs have been observed, with different MHV strains serving as prototypes of rodent CoVs that can infect variant tissues, with the MHV-A59 strain being predominantly hepatophilic and the MHV-JHM strain neurotropic [
30,
31,
32,
33]. Rodent coronavirus (RCoV) and sialodacryoadenitis virus (SDAV) both primarily infect the respiratory tract [
34]. In the present study, highly similar strains to HKU24 were detected in
A. chevrieri,
A. ilex and
R. norvegicus, which again suggested that the A lineage of β-CoV had intestinal tropics. Another cluster of α-CoVs: Poland
Myodes glareolus 1 (PLMg1), United Kingdom
Microtus agrestis 1 (UKMa1), United Kingdom
Microtus agrestis 2 (UKMa2) and United Kingdom
Rattus norvegicus 1 (UKRn1) in one of the lineages of α-CoV were only detected in liver samples of Norway rats, the bank vole, the wood mouse, and the noncyclic field vole, suggesting that they are hepatotropic [
19]. In the last 30 years, a number of cross-species transmission events of CoVs, as well as changes in viral tropism, have led to major new animal and human diseases involving bovine coronavirus (BCoV), HCoV-OC43, HCoV-229E, canine coronavirus (CCoV), feline coronavirus (FCoV), porcine coronavirus (PCoV), transmissible gastroenteritis virus (TGEV), and also the recently emerged severe SARS-CoV-2 [
35,
36,
37,
38,
39,
40,
41,
42]. While SARS-CoV-2 has emerged as the most recent example of zoonotic virus spillover to humans, studies have shown that SARS-CoV-2 also has widespread tissue tropism [
36]. The ability of CoV to cross species barriers and gradually spread to host animals in close contact with humans highlights the need to characterize small mammalian infections with coronaviruses, and it is also unlikely that SARS-CoV-2 will be the last CoV to cross species barriers and infect humans and other animal species [
43].
Viral surveillance in animal reservoirs is an important step to understand the exposure of humans to potential zoonoses, the types of human-animal interaction that impact the potential for spillover infection and the factors that determine the transmissibility and pathogenicity of viral zoonoses in humans [
44]. Then, whether small mammals infected with CoVs have any impact on human health and life remains to be further studied. Our current understanding of the diversity of viruses carried by small mammals, the host range of viruses, the drivers and specific mechanisms of cross-species transmission of viruses to humans is still shallow, limiting our indepth study of pathogens, and thus research in this area needs to be strengthened and requires special attention.
5. Conclusions
In this study, we collected a total of 502 small mammals belonging to 18 species in 12 genera and 4 orders collected in Dali and Nujiang Prefecture of Western Yunnan Province, China. The total number of CoV positives in the intestinal tissue samples was 20, including β-CoV (13) and α-CoV (7), with a prevalence of 3.98%.The prevalence of β-CoV was 3.54% (4/113) and 6.67% (6/90) in Apodemus chevrieri and A. ilex, respectively. The co-infection rates of both β-CoV and α-CoV in Eothenomys cachinus and Rattus norvegicus was 2.22% (2/90), 3.85% (1/26), respectively. The prevalence of α-CoV in R. nitidus and Episoriculus leucops was 75% (3/4), and 5.88% (1/17), respectively.
The qRT-PCR method based on a Taqman probe was designed for the detection of α-CoV and β-CoV in small mammalian samples with good sensitivity, specificity, stability and reproducibility. It was found that the detection rate of qRT-PCR was significantly higher than that of RT-PCR (P < 0.01). And by this method, we found that intestinal tissue contained the highest number of CoV copies (P < 0.0001) in the detected tissues. Establishing this method not only enables rapid epidemiological investigation but also helps to provide scientific support for the study of the epidemiology and pathogenesis of CoV infected by small mammals.
Supplementary Materials
Tables S1 and S2 partial RdRp nucleotide and amino acid sequence identity alignment of α-CoV and β-CoV, respectively. Tables S3 and S4 intra-group and inter-group repeatability test, respectively.
Author Contributions
Conceptualization, F.-H.X. and Y.-Z.Z.; methodology, F.-H.X., J.-W.T., X.-Y.G. and Y.-Z.Z.; software, P.-Y.H., Z.Y. and H.-M.Y.; validation, F.-H.X., Z.Y., P.-Y.H., L.-D.Z., H.-M.Y. and Y.-Z.Z.; formal analysis, F.-H.X., J.-Y.Z., W.K. and P.-Y.H.; investigation, H.-M.Y., L.-D.Z., Z.Y., J.-Y.Z. and W.K.; resources, P.-Y.H., J.-W.T., J.-Y.Z., W.K. and Y.-Z.Z.; data curation, F.-H.X., P.-Y.H. and Y.-Z.Z.; writing—original draft preparation, F.-H.X. and Y.-Z.Z.; writing—review and editing, F.-H.X., J.-W.T., P.-Y.H., X.-Y.G. and Y.-Z.Z.; supervision, H.-M.Y., L.-D.Z., X.-Y.G. and Y.-Z.Z.; project administration, F.-H.X. and Y.-Z.Z.; funding acquisition, Y.-Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (No. U2002218, 81874274); Yunnan Health Training Project of High Level Talents (No. L-2017027); Doctoral Research Start-up Fee Project of Dali University (No. KYBS2018004); Fund of Hunan University (521119400156); Co-funded Project of Cross-border Control and Quarantine Innovation Group of Zoonosis of Dali University (No. ZKPY2019302).
Data Availability Statement
The dataset analyzed during the current study are available from the corresponding authors on reasonable request. All the sequences in this manuscript can be obtained from NCBI database (
https://www.ncbi.nlm.nih.gov).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhou, Z.; Qiu, Y.; Ge, X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim Dis 2021, 1. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Yip, C.C.; Huang, Y.; Tsoi, H.W.; Chan, K.H.; Yuen, K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol 2006, 80, 7136–7145. [Google Scholar] [CrossRef] [PubMed]
- Hamre, D.; Procknow, J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 1966, 121, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Tsang, A.K.; Fan, R.Y.; Luk, H.K.; Cai, J.P.; Chan, K.H.; Zheng, B.J.; Wang, M.; et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol 2015, 89, 3076–3092. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Song, H.D.; Tu, C.C.; Zhang, G.W.; Wang, S.Y.; Zheng, K.; Lei, L.C.; Chen, Q.X.; Gao, Y.W.; Zhou, H.Q.; Xiang, H.; et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zeng, L.P.; Yang, X.L.; Ge, X.Y.; Zhang, W.; Li, B.; Xie, J.Z.; Shen, X.R.; Zhang, Y.Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Kin, N.; Miszczak, F.; Diancourt, L.; Caro, V.; Moutou, F.; Vabret, A.; Ar Gouilh, M. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect Genet Evol 2016, 40, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays J Pathol 2020, 42, 3–11. [Google Scholar] [PubMed]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Cheever, F.S.; Daniels, J.B. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. J Exp Med 1949, 90, 181–210. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.C.; Cross, S.S.; Rowe, W.P. Rat coronavirus (RCV): A prevalent, naturally occurring pneumotropic virus of rats. Arch Gesamte Virusforsch 1970, 31, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, X.D.; Guo, W.P.; Zhou, R.H.; Wang, M.R.; Wang, C.Q.; Ge, S.; Mei, S.H.; Li, M.H.; Shi, M.; et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 2015, 474, 19–27. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Wasberg, A.; Raghwani, J.; Li, J.; Pettersson, J.H.; Lindahl, J.F.; Lundkvist, A.; Ling, J. Discovery of a Novel Coronavirus in Swedish Bank Voles (Myodes glareolus). Viruses 2022, 14, 1205. [Google Scholar] [CrossRef]
- Tsoleridis, T.; Onianwa, O.; Horncastle, E.; Dayman, E.; Zhu, M.; Danjittrong, T.; Wachtl, M.; Behnke, J.M.; Chapman, S.; Strong, V.; et al. Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses 2016, 8, 84. [Google Scholar] [CrossRef]
- Lwande, O.W.; Mohamed, N.; Bucht, G.; Ahlm, C.; Olsson, G.; Evander, M. Seewis hantavirus in common shrew (Sorex araneus) in Sweden. Virol J 2020, 17, 198. [Google Scholar] [CrossRef]
- Ge, X.Y.; Yang, W.H.; Zhou, J.H.; Li, B.; Zhang, W.; Shi, Z.L.; Zhang, Y.Z. Detection of alpha- and betacoronaviruses in rodents from Yunnan, China. Virol J 2017, 14, 98. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Servent, A.; Francis, C.M. A new species of bat of the Hipposideros bicolor group (Chiroptera: Hipposideridae) from Central Laos, with evidence of convergent evolution with Sundaic taxa. Acta Chiropterologica 2006, 8, 39–61. [Google Scholar] [CrossRef]
- Watanabe, S.; Masangkay, J.S.; Nagata, N.; Morikawa, S.; Mizutani, T.; Fukushi, S.; Alviola, P.; Omatsu, T.; Ueda, N.; Iha, K.; et al. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg Infect Dis 2010, 16, 1217–1223. [Google Scholar] [CrossRef]
- Zhu, J.-H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y.-L.; Huang, Y.-W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J. Development and Clinical Applications of a 5-Plex Real-Time RT-PCR for Swine Enteric Coronaviruses. Viruses 2022, 14, 1536. [Google Scholar] [CrossRef]
- Hemida, M.G.; Chu, D.K.; Poon, L.L.; Perera, R.A.; Alhammadi, M.A.; Ng, H.Y.; Siu, L.Y.; Guan, Y.; Alnaeem, A.; Peiris, M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis 2014, 20, 1231–1234. [Google Scholar] [CrossRef]
- Hemida, M.G.; Elmoslemany, A.; Al-Hizab, F.; Alnaeem, A.; Almathen, F.; Faye, B.; Chu, D.K.; Perera, R.A.; Peiris, M. Dromedary Camels and the Transmission of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Transbound Emerg Dis 2017, 64, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, P.; Kolodziejek, J.; Khafaga, T.; Loney, T.; Howarth, B.; Sher Shah, M.; Abou Tayoun, A.; Alsheikh-Ali, A.; Camp, J.V.; Nowotny, N. Potentially Zoonotic Viruses in Wild Rodents, United Arab Emirates, 2019-A Pilot Study. Viruses 2023, 15, 695. [Google Scholar] [CrossRef]
- Chidoti, V.; De Nys, H.; Pinarello, V.; Mashura, G.; Missé, D.; Guerrini, L.; Pfukenyi, D.; Cappelle, J.; Chiweshe, N.; Ayouba, A.; et al. Longitudinal Survey of Coronavirus Circulation and Diversity in Insectivorous Bat Colonies in Zimbabwe. Viruses 2022, 14, 781. [Google Scholar] [CrossRef] [PubMed]
- Latif, A.A.; Mukaratirwa, S. Zoonotic origins and animal hosts of coronaviruses causing human disease pandemics: A review. Onderstepoort J Vet Res 2020, 87, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Hosking, M.P.; Lane, T.E. The pathogenesis of murine coronavirus infection of the central nervous system. Critical reviews in immunology vol 2010, 30, 119–130. [Google Scholar]
- Weiner, L.P. Pathogenesis of demyelination induced by a mouse hepatitis. Archives of neurology 1973, 28, 298–303. [Google Scholar] [CrossRef] [PubMed]
- De Albuquerque, N.; Baig, E.; Ma, X.; Zhang, J.; He, W.; Rowe, A.; Habal, M.; Liu, M.; Shalev, I.; Downey, G.P.; et al. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J Virol 2006, 80, 10382–10394. [Google Scholar] [CrossRef]
- Funk, C.J.; Manzer, R.; Miura, T.A.; Groshong, S.D.; Ito, Y.; Travanty, E.A.; Leete, J.; Holmes, K.V.; Mason, R.J. Rat respiratory coronavirus infection: Replication in airway and alveolar epithelial cells and the innate immune response. J Gen Virol 2009, 90, 2956–2964. [Google Scholar] [CrossRef]
- Liang, S.C.; Schoeb, T.R.; Davis, J.K.; Simecka, J.W.; Cassell, G.H.; Lindsey, J.R. Comparative Severity of Respiratory Lesions of Sialodacryoadenitis Virus and Sendai Virus Infections in LEW and F344 Rats. Vet Pathol 1995, 32, 661–667. [Google Scholar]
- Alekseev, K.P.; Vlasova, A.N.; Jung, K.; Hasoksuz, M.; Zhang, X.; Halpin, R.; Wang, S.; Ghedin, E.; Spiro, D.; Saif, L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol 2008, 82, 12422–12431. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cebra, C.K.; Baker, R.J.; Mattson, D.E.; Cohen, S.A.; Alvarado, D.E.; Rohrmann, G.F. Analysis of the genome sequence of an alpaca coronavirus. Virology 2007, 365, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Desario, C.; Mari, V.; Campolo, M.; Lorusso, E.; Elia, G.; Martella, V.; Buonavoglia, C.; Decaro, N. Molecular characterization of a canine respiratory coronavirus strain detected in Italy. Virus Res 2009, 141, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat Rev Microbiol 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Pfefferle, S.; Oppong, S.; Drexler, J.F.; Gloza-Rausch, F.; Ipsen, A.; Seebens, A.; Müller, M.A.; Annan, A.; Vallo, P.; Adu-Sarkodie, Y.; et al. Distant Relatives of Severe Acute Respiratory Syndrome Coronavirus and Close Relatives of Human Coronavirus 229E in Bats, Ghana. Emerg Infect Dis 2009, 15, 1377–1384. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol 2006, 80, 7270–7274. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Moes, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 2005, 79, 1595–1604. [Google Scholar] [CrossRef]
- Goraichuk, I.V.; Arefiev, V.; Stegniy, B.T.; Gerilovych, A.P. Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2. Virus Res 2021, 302. [Google Scholar] [CrossRef] [PubMed]
- Berto, A.; Anh, P.H.; Carrique-Mas, J.J.; Simmonds, P.; Van Cuong, N.; Tue, N.T.; Van Dung, N.; Woolhouse, M.E.; Smith, I.; Marsh, G.A.; et al. Detection of potentially novel paramyxovirus and coronavirus viral RNA in bats and rats in the Mekong Delta region of southern Viet Nam. Zoonoses Public Health 2018, 65, 30–42. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Sampling location. The left figure is a map of China, the right figure is a map of Yunnan Province, and the red dots in the figure represent the sampling points of this study.
Figure 1.
Sampling location. The left figure is a map of China, the right figure is a map of Yunnan Province, and the red dots in the figure represent the sampling points of this study.
Figure 2.
Co-evolution between coronaviruses and their hosts. The left is the phylogenetic tree established by CoV RdRp fragments, in which the virus strain marked in red is the sequence obtained in this study, and the hosts other than Muridae, Cricetidae, and Soricidae were labeled; the right is a phylogenetic tree constructed from the host mt-Cytb gene.
Figure 2.
Co-evolution between coronaviruses and their hosts. The left is the phylogenetic tree established by CoV RdRp fragments, in which the virus strain marked in red is the sequence obtained in this study, and the hosts other than Muridae, Cricetidae, and Soricidae were labeled; the right is a phylogenetic tree constructed from the host mt-Cytb gene.
Figure 3.
Standard curve. (A) The equation of the standard curve of α-CoV is y = −3.2297x + 43.576, the correlation coefficient R2 = 0.9974, the slope = −3.2297, and the amplification efficiency (E%) = 104%. (B) The equation of the standard curve of β-CoV is y = −3.4586x + 44.401, the correlation coefficient R2 = 0.9991, the slope = −3.4586, the amplification efficiency (E%) = 95%.
Figure 3.
Standard curve. (A) The equation of the standard curve of α-CoV is y = −3.2297x + 43.576, the correlation coefficient R2 = 0.9974, the slope = −3.2297, and the amplification efficiency (E%) = 104%. (B) The equation of the standard curve of β-CoV is y = −3.4586x + 44.401, the correlation coefficient R2 = 0.9991, the slope = −3.4586, the amplification efficiency (E%) = 95%.
Figure 4.
Sensitivity test results. (A) α-CoV. (B) β-CoV. 9–1: Copies: 1.00 × 109–1.00 × 101 copies/µL; 0: Negative control.
Figure 4.
Sensitivity test results. (A) α-CoV. (B) β-CoV. 9–1: Copies: 1.00 × 109–1.00 × 101 copies/µL; 0: Negative control.
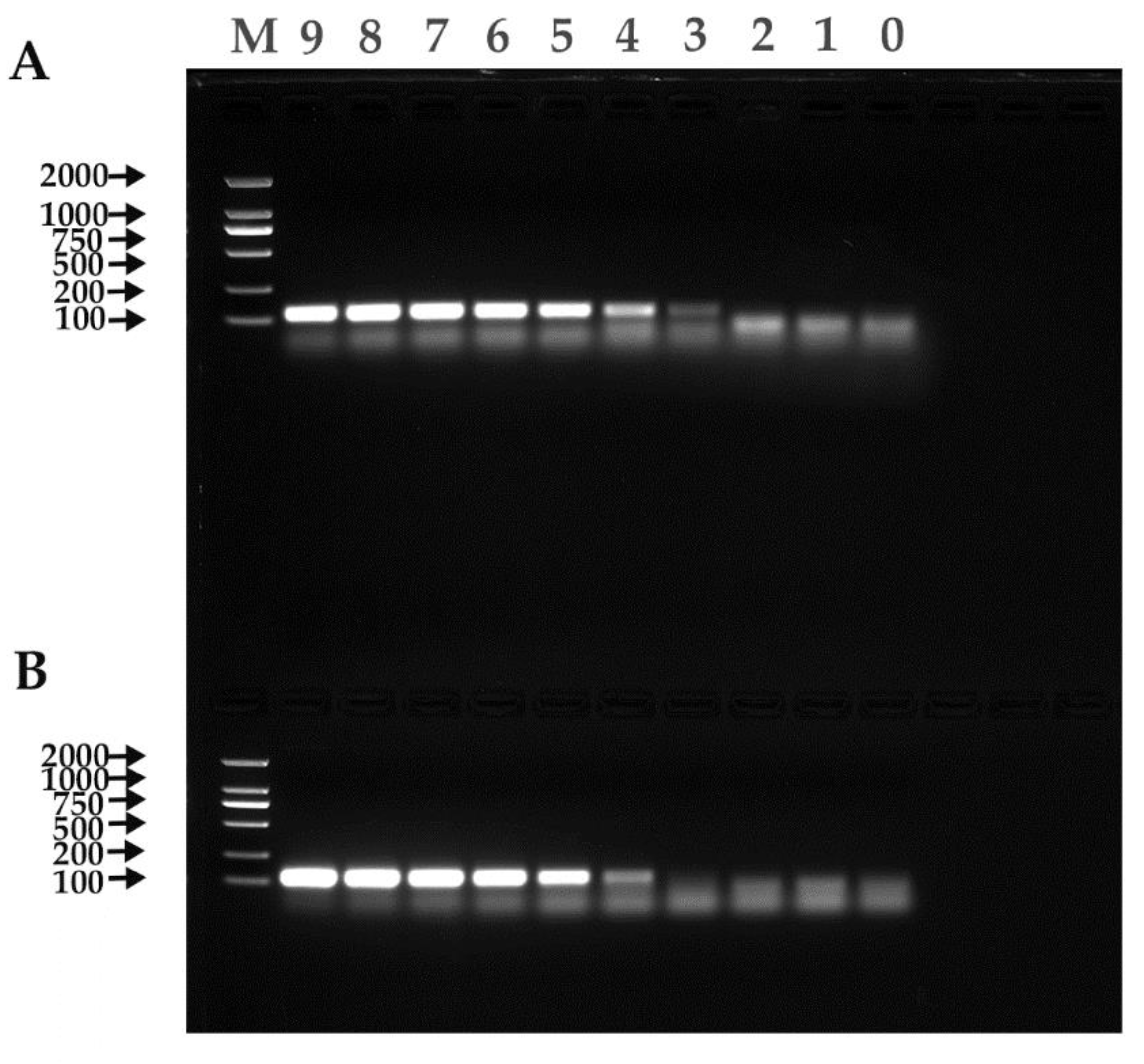
Figure 5.
RT-PCR results. (A) α-CoV. (B) β-CoV. M: Trans2K DNA Marker; 9–1: 1.00 × 109–1.00 × 101 copies/µL; 0: Negative control.
Figure 5.
RT-PCR results. (A) α-CoV. (B) β-CoV. M: Trans2K DNA Marker; 9–1: 1.00 × 109–1.00 × 101 copies/µL; 0: Negative control.
Figure 6.
Tissue distribution of CoV in positive samples. The quantification (mean ± standard error) of CoV RNA in the intestine, liver, heart, spleen, lung and kidney of 20 CoV-positive samples was measured in copies/µL, and the significance test was performed using one-way ANOVA (****P < 0.0001).
Figure 6.
Tissue distribution of CoV in positive samples. The quantification (mean ± standard error) of CoV RNA in the intestine, liver, heart, spleen, lung and kidney of 20 CoV-positive samples was measured in copies/µL, and the significance test was performed using one-way ANOVA (****P < 0.0001).
Table 1.
The primer information in this study.
Table 1.
The primer information in this study.
| |
Primer name |
Sequence (5′ → 3′) |
bp |
Amplify the region |
References |
CoV
RT-PCR primer |
CoV-FWD3 |
GGTTGGGAYTAYCCHAARTGTGA |
434 bp |
RdRp |
[23] |
| CoV-FWD4/other |
GAYTAYCCHAARTGTGAUMGWGC |
| CoV-RVS3 |
CCATCATCASWYRAATCATCATA |
α-RCoV
qRT-PCR primer and probe |
α-RCoV-F14493 |
ACATCTGGTGATGCTAGTAC |
110 bp |
RdRp |
This study |
| α-RCoV-R14602 |
TTCCTRCAAACATTACTATCAACAG |
| α-RCoV-Probe |
FAM-TTTTCAGGCTGTTAGTGCTAATGTAAATAAATTGC-BHQ1 |
β-RCoV
qRT-PCR primer and probe |
β-RCoV-F15324 |
AGTATGATGATTTTGAGTGATGATGGYGTTG |
117 bp |
RdRp |
This study |
| β-RCoV-R15440 |
CACGTTATTTTGATAATACAGCACCTGTTG |
| β-RCoV-Probe |
FAM-TATGCGTCCAAAGGTTATATTGCTAATATTAGTGCCT-BHQ1 |
| Rodent identification primer |
L14724 |
ATGATATGAAAAACCATCGTTG |
1200 bp |
mt-Cytb
|
[22] |
| H15915 |
TTTCCNTTTCTGGTTTACAAGAC |
Table 2.
The situation of CoV infection in small mammals in Dali and Nujiang, Yunnan Province.
Table 2.
The situation of CoV infection in small mammals in Dali and Nujiang, Yunnan Province.
| Order |
Species |
Locations |
Composition, % |
Prevalence, % |
| qRT-PCR |
RT-PCR |
|
| α-CoV |
β-CoV |
| Rodentia |
Apodemus chevrieri |
Dali, Nujiang |
22.51 (113/502) |
9.73 (11/113) |
0 (0/113) |
3.54 (4/113) |
| Apodemus ilex |
Dali, Nujiang |
17.93 (90/502) |
21.11 (19/90) |
0 (0/90) |
6.67 (6/90) |
| Eothenomys cachinus |
Dali, Nujiang |
16.14 (81/502) |
11.11 (9/81) |
2.22 (2/90) |
2.22 (2/90) |
| Eothenomys miletus |
Dali, Nujiang |
7.77 (39/502) |
12.82 (5/39) |
0 (0/39) |
0 (0/39) |
| Rattus tanezumi |
Dali, Nujiang |
14.94 (75/502) |
25.33 (19/75) |
0 (0/75) |
0 (0/75) |
| Rattus norvegicus |
Dali |
5.18 (26/502) |
46.15 (12/26) |
3.85 (1/26) |
3.85 (1/26) |
| Niviventer eha |
Nujiang |
0.6 (3/502) |
0 (0/3) |
0 (0/3) |
0 (0/3) |
| Mus caroli |
Dali |
0.6 (3/502) |
0 (0/3) |
0 (0/3) |
0 (0/3) |
| Mus musculus |
Dali |
0.40 (2/502) |
0 (0/2) |
0 (0/2) |
0 (0/2) |
| Niviventer fulvescens |
Dali, Nujiang |
0.40 (2/502) |
0 (0/2) |
0 (0/2) |
0 (0/2) |
| Rattus nitidus |
Dali |
0.80 (4/502) |
75 (3/4) |
75 (3/4) |
0 (0/4) |
| Tamiops swinhoei |
Dali |
0.20 (1/502) |
0 (0/1) |
0 (0/1) |
0 (0/1) |
| Insectivora |
Blarina brevicauda |
Nujiang |
4.18 (21/502) |
4.67 (1/21) |
0 (0/21) |
0 (0/21) |
| Episoriculus leucops |
Nujiang |
3.39 (17/502) |
5.88 (1/17) |
5.88 (1/17) |
0 (0/17) |
| Crocidura attenuata |
Nujiang |
2.19 (11/502) |
27.27 (3/11) |
0 (0/11) |
0 (0/11) |
| Suncus murinus |
Dali |
0.60 (3/502) |
66.67 (2/3) |
0 (0/3) |
0 (0/3) |
| Lagomorpha |
Ochotona thibetana |
Nujiang |
1.79 (9/502) |
11.11 (1/9) |
0 (0/9) |
0 (0/9) |
| Scandentia |
Tupaia belangeri |
Dali |
0.40 (2/502) |
50 (1/2) |
0 (0/2) |
0 (0/2) |
| |
Total |
|
100 (502/502) |
17.33 (87/502) |
1.39(7/502) |
2.59 (13/502) |
Table 3.
CoV copies in tissue of positive samples.
Table 3.
CoV copies in tissue of positive samples.
| |
Intestinal |
Liver |
Heart |
Spleen |
Lung |
Kidney |
| Mean (copies/g) |
1.25 × 106
|
4.02 × 103
|
2.89 × 103
|
2.88 × 103
|
1.68 × 103
|
1.29 × 103
|
| SEM |
1.09 × 106
|
2.71 × 103
|
2.28 × 103
|
2.77 × 103
|
1.47 × 103
|
1.09 × 103
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).