1. Introduction
Intrinsically disordered proteins (IDPs) represent a fascinating class of proteins that lack well-defined three-dimensional structures under physiological conditions [
1,
2]. Despite their apparent structural disorder, IDPs play crucial roles in various cellular processes and are implicated in numerous diseases [
3,
4,
5].
In recent decades, the study of IDPs has substantially expanded our understanding of protein structure-function relationships, challenging the traditional paradigm of "one structure, one function." Scientists have unveiled the remarkable ability of IDPs to fold their intrinsically disordered segments into functionally active domains [
6]. These disordered sequences can fluctuate in form from collapsed globules to statistical coils, yet they are still able to effectively bind biological targets [
7]. The dynamic nature of IDPs enables one singular protein to interact with a range of biological targets, challenging the traditional view that protein function is tied to a strict three-dimensional structure [
7]. However, the broader scientific community has not fully embraced IDPs. Many researchers still adhering to the classical structure-function dogma and often times dismissing the importance and relevance of IDPs. Therefore, it is imperative to utilize available tools to the community that can help educate the scientific community about IDPs, their physiological importance, and their role in human diseases.
In the fast-paced landscape of technological innovation, artificial intelligence (AI) has carved a significant niche for itself, becoming an influential player across myriad applications, notably in scientific research and pedagogical domains. A notable example of such AI-driven tools is the GPT-4 model, a product of OpenAI. This AI language model harnesses the power of internet-based data to forecast the most probable succeeding words in any given sentence, making it a powerful tool in understanding and generating human-like text [
8].
The GPT-4 model embodies the principles of Transformer-style architecture, a neural network designed to process and generate data in sequence. Its training paradigm involves predicting subsequent tokens (I.e., the basic units of text or code that an LLM AI uses to process and generate language) within a document. The vast corpus of training data hails from a variety of sources, encompassing publicly accessible data (e.g., internet-based data) and datasets obtained through licensing from third-party entities [
8]. To enhance its performance, GPT-4 has been fine-tuned using a technique known as Reinforcement Learning from Human Feedback (RLHF) [
8].
Known as a large language model (LLM), GPT-4 can deliver large volumes of information on a variety of elaborate topics to users in a palatable, conversational manner [
9]. One evaluation of GPT-3.5, a predecessor of GPT-4, found that the AI language model was able to score at or near the 60% pass mark for all three of the United States Medical Licensing Examinations (USMLEs), while medical students spend anywhere from 300-400 hours of focused study time to prepare for solely the first of these three examinations [
10,
11]. Further exploration of GPT-4's capabilities has shown its aptitude to tackle a broad range of intricate prompts across disciplines such as mathematics, coding, vision, law, and psychology while maintaining a human-like output [
9,
12]. These analyses of ChatGPT demonstrate the power of AI tools to further understand complex topics, including IDPs. As such, we posit that harnessing the power of AI tools like LLMs could significantly enhance the comprehension and recognition of IDPs within the scientific community.
In this study, we present an empirical analysis that assesses the performance of several LLMs, including GPT-4, GPT-3.5, Google Bard (PaLM 2), GPT-4, and GPT-4 with Bing Browser plugin, in the context of education on Intrinsically Disordered Proteins (IDPs). The analysis is based on a dataset gathered from a survey and focuses on five evaluation categories: Accuracy, Relevance, Depth of Understanding, Clarity, and Overall Quality.
As part of the evaluation, we engage in a dialogue with the AI models to explore various facets of IDPs, including common misconceptions, challenges in research, and the future trajectory of the field. This dialogue helps in analyzing the capacity of these AI models to disseminate knowledge on IDPs and foster an inclusive understanding of protein structure-function relationships. To further bolster our analysis, we have enlisted the expertise of Dr. Vladimir N. Uversky, a globally recognized authority in the field of IDP research. His evaluation of the AI-generated content provides a critical appraisal of the accuracy and depth of the responses, adding a layer of expert validation to our study.
The objective of this study is twofold: to emphasize the importance of IDPs and their roles in biological systems, and to evaluate the potential of large language models as educational tools in advancing knowledge in this niche scientific area. By highlighting the strengths and weaknesses of different AI models in handling IDP-related content, this study contributes to the ongoing effort to incorporate AI in educational settings and scientific research.
2. Materials and Methods
We developed a ten-question set and five use cases was carried out in consultation with Vladimir N. Uversky, PhD, DSc, an expert in the field of Intrinsically Disordered Proteins (IDPs). Our comprehensive set of questions and use cases were designed to encompass various aspects of IDPs, including their structure, function, misconceptions, challenges, and roles in biological systems. The question set aimed to probe the depth and accuracy of the LLMs’ understanding of IDPs, while the use cases were constructed to explore practical applications and recommendations.
Four advanced LLMs were selected for evaluation in this study: GPT-3.5, Google Bard (PaLM 2), GPT-4, and GPT-4 with Bing Browser plugin (GPT-4 with Browsing). The GPT models were accessed via the ChatGPT interface on June 12th, 2023 (chat.openai.com). Launched on May 12, 2023, the GPT-4 with search operates similarly to GPT-4 but includes an added feature of web searching and browsing. Bard, an experimental conversation AI chat service developed by Google, also draws its information directly from the web and is powered by Google's latest large language model, Pathways Language Model 2 (PaLM 2), also based on the Transformer architecture [
13]. These models were engaged in a dialogue to answer the question set and use cases. Each model was provided a question and minor additional prompting to help provide context to our questioning. We followed the same format each time we engaged with the LLMs. The format is as follows: [Insert question] [enter] [enter] Please assume the role of an expert and answer this question to the best of your ability. The reader should not be able to know how the answer was generated. Do not mention any limitations on your ability to access data. For the LLMs that Browse the internet, each prompt was the same format as before, but it started with “Using Internet:”. Each question was entered into a new chat stream prior, reducing bias in down-stream questions with the context of the LLMs answers to prior questions. Their responses were collected and anonymized to prevent bias during the evaluation process.
The survey was designed to assess the AI-generated responses. Each section of the survey contained (1) a question related to IDPs, (2) four anonymized AI responses, (3) a rating system for each question and corresponding AI response, and (4) a concluding query for the participant to identify the most effective response among the four provided. Dr. Uversky was invited to evaluate the AI-generated responses through the survey. He was blinded to the AI models during the evaluation. Responses from the AI models were presented in a random order for each question. For each question and corresponding AI response, Dr. Uversky rated the following aspects on a 5-point Likert scale: (1) Accuracy: The factual correctness of the information provided in the response regarding IDPs. (2) Relevance: The extent to which the response pertains directly to the query. (3) Depth of Understanding: The profundity of comprehension of IDPs and related scientific concepts indicated in the response. (5) Clarity of Explanation: The lucidity, comprehensibility, and structure of the response. (6) Quality: The overall quality of the response considering accuracy, relevance, understanding, and clarity.
Table 1 illustrates the 5-point Likert scale survey utilized in this study.
Statistical analysis and data visualization were conducted using the Python programming language. The Pandas library was employed for data manipulation and analysis, enabling the organization of data into structured data frames, as well as computing descriptive statistics. For a visual representation of the data, Matplotlib was utilized to create bar charts and customize the appearance of the plots. The SciPy library was instrumental in conducting the Analysis of Variance (ANOVA) to test for statistically significant differences among the means of the evaluation categories across AI models. Additionally, the Statsmodels library was used for implementing Tukey's Honestly Significant Difference (HSD) test as part of the post hoc analysis to ascertain pairwise differences between AI models in each evaluation category.
3. Results
Our study evaluates the performance of four AI models - GPT-3.5, GPT-4, GPT-4 with Browsing, and Google Bard (PaLM 2) - in the context of Intrinsically Disordered Proteins (IDPs). A comprehensive set of assessment questions and use cases (
Table 2) was employed to probe the AI models' knowledge and understanding of IDPs. The expert evaluation was provided by Dr. Vladimir N. Uversky, who was blinded to the AI models during the evaluation. His insights enriched the analysis and offered a critical perspective on the AI models' performance. Each AI model's responses are catalogued in
Supplementary Figure S1, with the expert-chosen preferred responses distinguished by a green highlight. An example of such a preferred response, generated by GPT-4, is demonstrated in
Figure 1.
Descriptive statistics were computed for each AI model across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. The mean and standard deviation of the performance ratings for each AI model were tabulated and are presented in
Table 3, offering insights into trends and variations in the AI models' performance. Graphical representation of the average performance ratings of the AI models in these categories is provided in
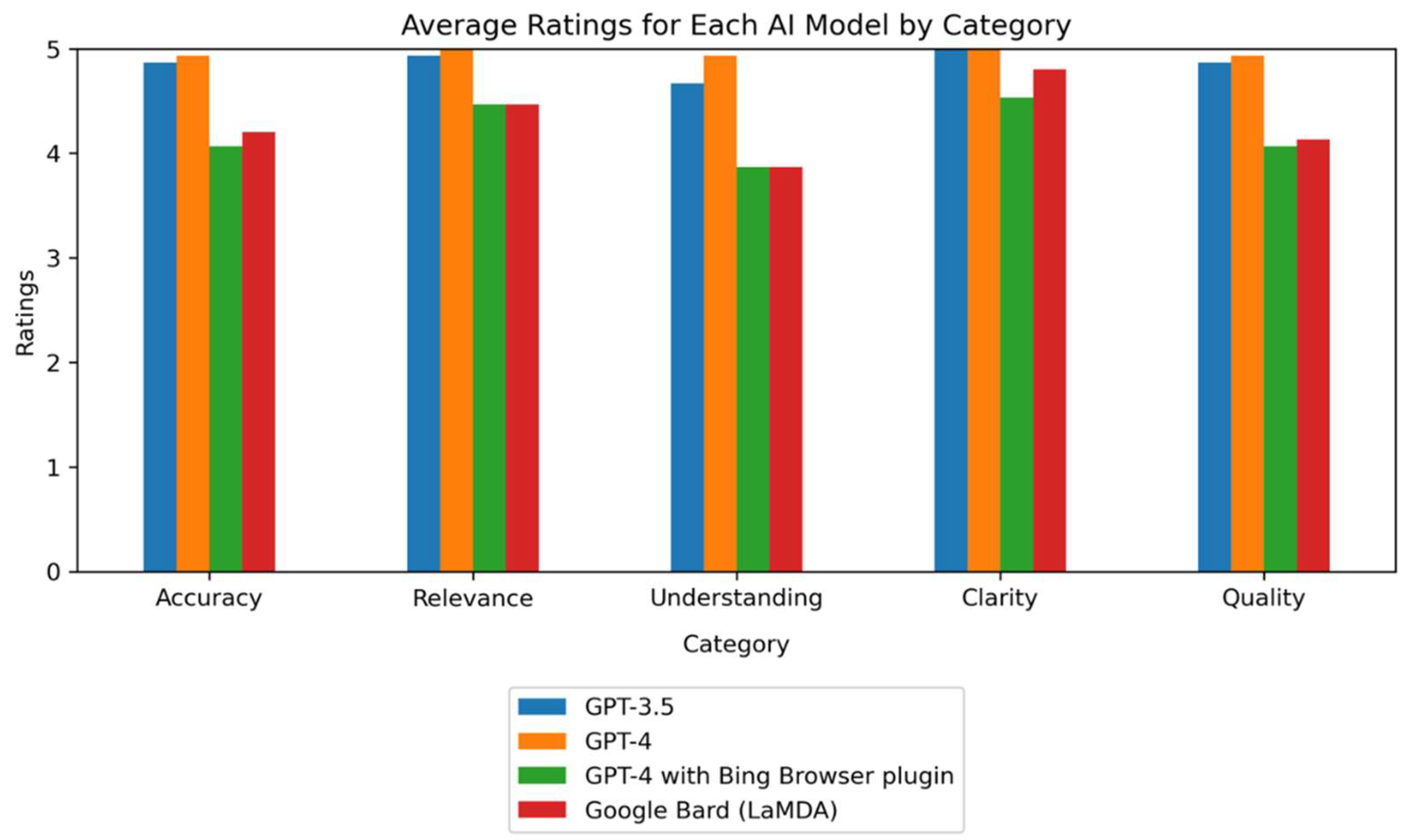
Figure 2, which visually represents how each AI model performed in the different evaluation categories.
An Analysis of Variance (ANOVA) was conducted to assess if there were any statistically significant differences among the means of the groups. The ANOVA analysis revealed significant differences between the AI models in terms of accuracy, relevance, understanding, clarity, and quality. The F statistics and p-values for each category were as follows: Accuracy (F=16.77, p<0.0001), Relevance (F=6.79, p=0.0006), Understanding (F=16.14, p<0.0001), Clarity (F=6.70, p=0.0006), and Quality (F=24.61, p<0.0001).
To discern which groups, have significant differences, a post hoc analysis using Tukey's Honestly Significant Difference (HSD) test was conducted. The results of the Tukey's HSD test are presented in
Table 4. This test helped in understanding the pairwise differences between the AI models in each evaluation category. The mean differences, confidence intervals, and decisions to reject or not reject the null hypothesis are reported for each pairwise comparison. In the Tukey's HSD test data for the "Accuracy" category, it is observed that GPT-4 with Browsing and Google Bard (PaLM 2) had significantly lower ratings compared to GPT-3.5 and GPT-4, while there were no significant differences between GPT-3.5 and GPT-4, or between GPT-4 with Browsing and Google Bard (PaLM 2). In the "Relevance" category, GPT-4 with Browsing and Google Bard (PaLM 2) were significantly lower than GPT-3.5, and GPT-4 with Browsing was significantly lower than GPT-4. For "Understanding," GPT-4 with Browsing and Google Bard (PaLM 2) were significantly lower than both GPT-3.5 and GPT-4. In the "Clarity" category, GPT-4 with Browsing was significantly lower than both GPT-3.5 and GPT-4. Lastly, for the "Quality" category, GPT-4 with Browsing and Google Bard (PaLM 2) were significantly lower than both GPT-3.5 and GPT-4.
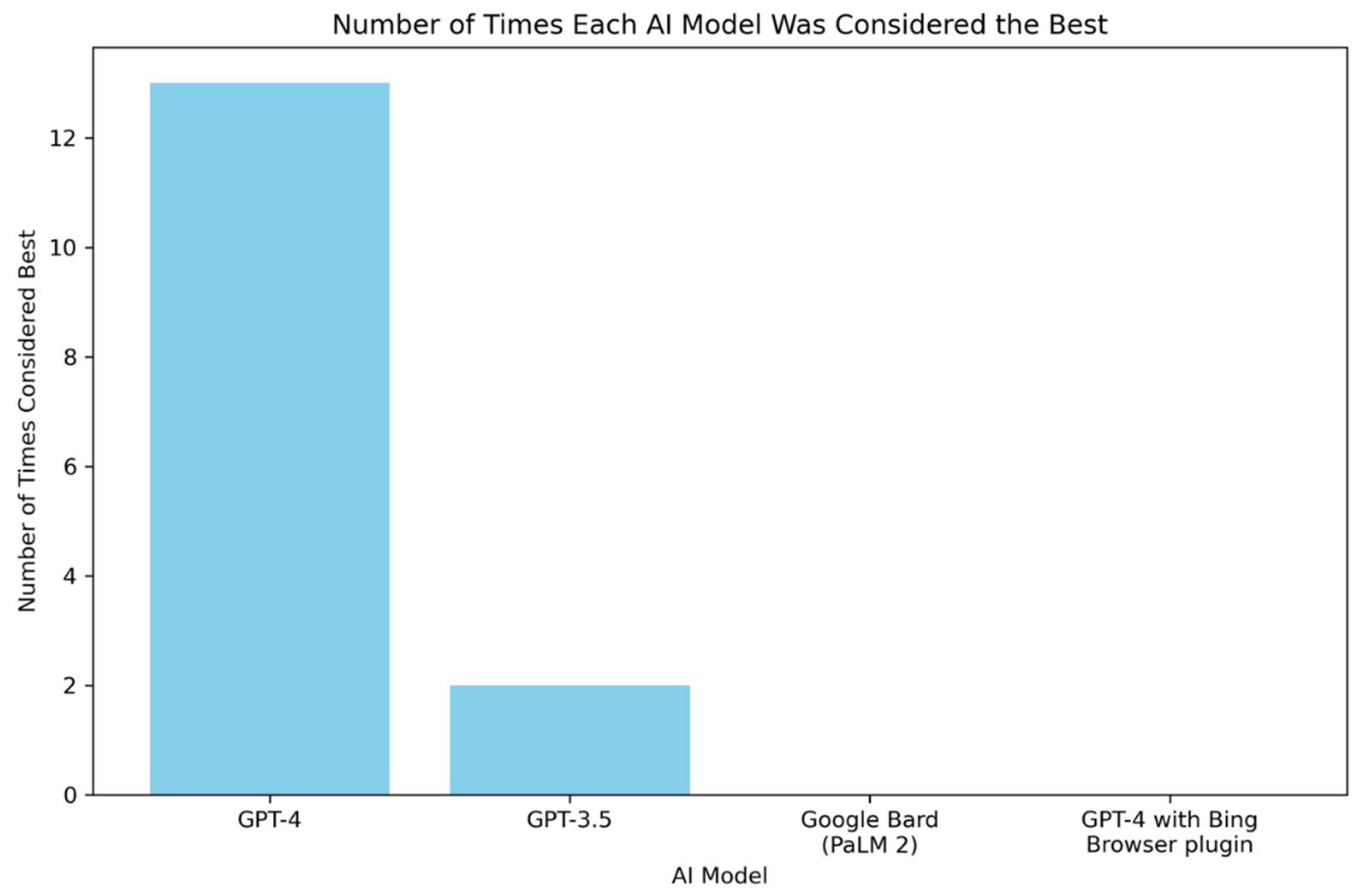
Additionally, a bar chart depicting the number of times each AI model was considered the best in handling questions related to IDPs is presented in
Figure 3. This chart demonstrates the frequency with which each AI model was acknowledged for its exemplary performance in the study. Notably, GPT-4 emerges as a clear favorite among the models, receiving 13 votes for being considered the best in handling questions related to IDPs. In stark contrast, GPT-3.5 garnered only 2 votes. Google Bard (PaLM 2) and GPT-4 with Browsing did not receive any votes, indicating a lack of preference for the answers generated by these LLMs.
4. Discussion
Our investigation encompassed an empirical evaluation of various AI language models, with a particular focus on their utility in the educational domain of Intrinsically Disordered Proteins (IDPs). The models assessed were GPT-3.5, GPT-4, GPT-4 with Browsing, and Google Bard (PaLM 2). The performance was evaluated across five categories: accuracy, relevance, depth of understanding, clarity, and overall quality.
One of the key findings of this study was the superior performance of GPT-4 in handling questions related to IDPs. GPT-4 demonstrated the highest mean ratings across all evaluation categories. Notably, it was also chosen as the most effective AI model in 13 out of 15 instances, far surpassing the performance of the other models.
We posit that the underlying architecture and extensive training data of GPT-4 may have contributed to its effectiveness. GPT-4.0 demonstrates improved abilities in generating complex language patterns and interpretation of abstract concepts compared to its predecessor GPT-3.5 [
14]. According to OpenAI’s technical report, GPT-4 was able to score in the 90
th percentile on a simulated Uniform Bar Exam, while GPT-3.5 scored at the 10
th percentile [
8]. GPT-4.0 also scored in the 88
th percentile on the Law School Admission Test (LSAT), while GPT-3.5 only scored in the 40
th percentile [
8]. With regards to scientific content, GPT-4.0 was able to score in the 85
th-100
th percentile on Advanced Placement (AP) Biology and Statistics exams, while GPT-3.5 only scored in the 62
nd to 85
th percentile and 40
th to 63
rd percentile on the AP Biology and Statistics exams, respectively. On the Ophthalmology Knowledge Assessment Program (OKAP) examination, GPT-4.0 significantly outshined GPT-3.5 (81% vs. 57%; p<0.001) [
15]. When compared to GPT-3.5, GPT-4.0 exceeds it in nearly all facets, providing exceptional human-like performance on widely administered academic and professional exams. In a dataset of 5,214 prompts, human evaluators preferred GPT-4.0’s responses over GPT-3.5 in 70.2% of them [
8]. When compared to its competitors, GPT-4.0 scored 71 out of a possible 100 in discerning provided facts apart from fiction, outshining Google Bard (PaLM 2) and Microsoft Bing AI’s lesser 65.25 out of 100[
14], also reflecting the results of our study.
Our findings highlight an intriguing paradox concerning the utility of search capabilities in LLMs. While intuitively, the ability to actively fetch recent data from the web might seem beneficial, our analysis demonstrated that models employing this feature, such as GPT-4 with Browsing and Google Bard (PaLM 2), scored lower across all evaluation criteria when compared to their search-incapable counterparts, GPT-3.5 and GPT-4.
Despite GPT-4 and GPT-4 with Browsing sharing the same underlying LLM, GPT-4 outperformed its counterpart in all criteria, even being the preferred choice among evaluators. The underlying rationale for this outcome is that GPT-4 and GPT-3.5, which are trained on billions of parameters, can generate complex, comprehensive responses based on the input query alone. In contrast, GPT-4 with Browsing must initiate a web search via the Bing Browser plugin, read and comprehend individual webpages, and if necessary, return to the search page to seek additional information.
An additional hindrance to the browsing models is the occasional occurrence of a "click failed" error during the search process. According to OpenAI, this error is a result of the plugin honoring the website's instructions to avoid crawling, encapsulated in the site's robots.txt file. This is in place to respect content creators and adhere to the web’s norms. However, it introduces an element of unpredictability to the model's ability to gather necessary information.
The constraints of the browsing capability reveal themselves through two primary limitations. First, the quality of the LLM's response is contingent upon the quality and depth of information available and accessible during its web search. The LLM's response is, in essence, a derivative of the webpages it peruses, which may not always provide comprehensive or accurate information. Second, the browsing LLM is also limited by its context window. As it reads through web pages, much of which can span dozens of pages, it can only retain a portion of this information within its context window. Thus, despite the ability to reference specific sources, which provides transparency and allows users to further explore the cited information, the browsing LLM's potential to craft a detailed response is impeded.
Google Bard (PaLM 2) exhibited similar limitations, suggesting that the integration of search capabilities into LLMs may universally hinder their performance, at least in their current iterations. An interesting point to consider here is the alleged proclivity of LLMs towards "hallucinations" or fabricating details. Some argue that incorporating search capabilities could reduce this tendency. However, our study indicates that while it may limit hallucinations, it also appears to undermine the LLM's ability to answer complex questions, leading to inferior performance across the board.
It is essential to interpret these findings in light of the inherent strengths and weaknesses of LLMs. Our study indicates that the addition of search capabilities to LLMs is not an unequivocal enhancement. Future research should aim to strike a balance between enhancing the breadth of knowledge accessible to the LLMs and maintaining their inherent ability to generate complex, coherent, and creative responses.
The implications of this study for IDP research and education are manifold. The results suggest that AI models, particularly GPT-4, could serve as valuable educational tools for disseminating IDP knowledge. By providing accurate and comprehensible information on IDPs, these AI models can supplement traditional educational resources and foster a more inclusive understanding of protein structure-function relationships. Scientists have already begun leveraging AI in this manner, training a ChatGPT-like system on a database of 143,508 proteins. Users can interact extensively with this model, inquiring about uploaded proteins to foster deeper insights into protein structure and function [
16]. The findings of our study also support the use of AI systems to advance knowledge of the complex nature of proteins, particularly IDPs.
While this study is one of the few to focus on the application of AI models in the context of IDPs, there is a growing body of literature that examines the role of AI in scientific education more broadly. One study gathered ChatGPT’s responses to prompts about education in science and compared them with an accepted doctrine on effective scientific teaching. They found strong alignment between the two sources, with key components overlapping the two [
17]. Our findings align with the consensus that AI models have the potential to facilitate scientific education, though their efficacy can vary based on the topic and the specific model used.
It is important to acknowledge the limitations of this study. The evaluation is based on a limited set of questions and the assessment of the AI models was done by a single expert. Future studies could benefit from a more diverse set of evaluators, and potentially from incorporating feedback from students or educators who might use these tools in a real-world educational setting. It is also important to mention the limitations of using ChatGPT and other AI models in this role. Previous studies have often cited inaccurate, incorrect, and unimportant information when applying ChatGPT to conduct data collection and research [
18,
19]. Previous accounts have also noted that ChatGPT can provide inaccurate citations, sometimes even referencing non-existent literature [
20,
21]. These are aspects of ChatGPT that significantly limit its practicality in the scientific setting. Additionally, we must be careful with overutilizing ChatGPT in educational systems. A previous study found that students who utilized ChatGPT were more likely to commit plagiarism than those who did not [
22]. On February 17, 2023, the University of Hong Kong was the first higher education institution to ban students from using ChatGPT and other AI tools for coursework [
23]. Educators worry that overutilization of these tools can result in overreliance and a corresponding lack of critical thinking. Similarly, we must be cautious when moving forward with these AI models in understanding IDPs and, ultimately, in the navigation of the entire scientific field. However, AI developers can use the insights from this study to refine their models, particularly in the context of scientific education.
5. Conclusions
In conclusion, our study emphasizes the significant role of AI language models, especially GPT-4, in enhancing our understanding of intrinsically disordered proteins. Its clear from our results that as we move forward, the synergy between AI specialists, educators, and researchers will be paramount. Together, there is a promising opportunity to elevate the realm of scientific education.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Question and Answer with LLMs.
Author Contributions
Conceptualization, D.T.G., and M.D.; methodology, D.T.G., M.D., and V.N.U.; validation, D.T.G., M.D., and V.N.U.; formal analysis, D.T.G., and M.D.; investigation, D.T.G., M.D., C.K., and V.N.U.; data curation, D.T.G., and M.D..; writing—original draft preparation, D.T.G., M.D., C.K., M.A., and G.R.G.; writing—review and editing, D.T.G., M.D., C.K., M.A., G.R.G., and V.N.U.; visualization, D.T.G., and M.D.; supervision, D.T.G., M.D., and V.N.U.; project administration, D.T.G., and M.D.. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Any supplementary materials related to this manuscript are also represented within the paper.
Acknowledgments
We extend our gratitude to Google and OpenAI for granting access to their cutting-edge AI tools. Special thanks to the IDP researchers for their relentless pursuit of knowledge and their audacity to chart new territories in protein science.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baul, U., et al., Sequence Effects on Size, Shape, and Structural Heterogeneity in Intrinsically Disordered Proteins. J Phys Chem B, 2019. 123(16): p. 3462-3474. [CrossRef]
- Trivedi, R. and H.A. Nagarajaram, Intrinsically Disordered Proteins: An Overview. Int J Mol Sci, 2022. 23(22). [CrossRef]
- Bondos, S.E., A.K. Dunker, and V.N. Uversky, Intrinsically disordered proteins play diverse roles in cell signaling. Cell Communication and Signaling, 2022. 20(1): p. 20. [CrossRef]
- Uversky, V.N., Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des, 2013. 19(23): p. 4191-213. [CrossRef]
- Uversky, V.N., Introduction to Intrinsically Disordered Proteins (IDPs). Chemical Reviews, 2014. 114(13): p. 6557-6560. [CrossRef]
- Schlessinger, A., et al., Protein disorder--a breakthrough invention of evolution? Curr Opin Struct Biol, 2011. 21(3): p. 412-8. [CrossRef]
- Dyson, H.J. and P.E. Wright, Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology, 2005. 6(3): p. 197-208. [CrossRef]
- OpenAI, GPT-4 Technical Report. ArXiv, 2023. arXiv:2303.08774.
- Bubeck, S., et al., Sparks of artificial general intelligence: Early experiments with gpt-4. arXiv preprint arXiv:2303.12712, 2023. arXiv:2303.12712.
- Burk-Rafel, J., S.A. Santen, and J. Purkiss, Study Behaviors and USMLE Step 1 Performance: Implications of a Student Self-Directed Parallel Curriculum. Acad Med, 2017. 92(11S Association of American Medical Colleges Learn Serve Lead: Proceedings of the 56th Annual Research in Medical Education Sessions): p. S67-s74. [CrossRef]
- Kung, T.H., et al., Performance of ChatGPT on USMLE: Potential for AI-assisted medical education using large language models. PLOS Digit Health, 2023. 2(2): p. e0000198. [CrossRef]
- Rahman, M.M. and Y. Watanobe, ChatGPT for Education and Research: Opportunities, Threats, and Strategies. Applied Sciences, 2023. 13(9): p. 5783. [CrossRef]
- Anil, R., et al., Palm 2 technical report. arXiv preprint arXiv:2305.10403, 2023. [CrossRef]
- Caramancion, K., News Verifiers Showdown: A Comparative Performance Evaluation of ChatGPT 3.5, ChatGPT 4.0, Bing AI, and Bard in News Fact-Checking. arXiv pre-print server, 2023. [CrossRef]
- Teebagy, S., et al., Improved Performance of ChatGPT-4 on the OKAP Exam: A Comparative Study with ChatGPT-3.5. medRxiv, 2023: p. 2023.04.03.23287957. [CrossRef]
- Guo, H., et al., ProteinChat: Towards Achieving ChatGPT-Like Functionalities on Protein 3D Structures. 2023, TechRxiv.
- Cooper, G., Examining Science Education in ChatGPT: An Exploratory Study of Generative Artificial Intelligence. Journal of Science Education and Technology, 2023. 32(3): p. 444-452. [CrossRef]
- Stokel-Walker, C. and R. Van Noorden, What ChatGPT and generative AI mean for science. Nature, 2023. 614(7947): p. 214-216. [CrossRef]
- Moons, P. and L. Van Bulck, ChatGPT: can artificial intelligence language models be of value for cardiovascular nurses and allied health professionals. European Journal of Cardiovascular Nursing, 2023. [CrossRef]
- Chen, T.J., ChatGPT and other artificial intelligence applications speed up scientific writing. J Chin Med Assoc, 2023. 86(4): p. 351-353. [CrossRef]
- Lubowitz, J.H., ChatGPT, An Artificial Intelligence Chatbot, Is Impacting Medical Literature. Arthroscopy, 2023. 39(5): p. 1121-1122. [CrossRef]
- Bašić, Ž., et al., Better by You, better than Me? ChatGPT-3 as writing assistance in students’ essays. 2023. [CrossRef]
- Holliday, I. About ChatGPT. 2023 [cited 2023 July 23]; Available from: https://tl.hku.hk/2023/02/about-chatgpt/.
Figure 1.
Preferred Response Example from GPT-4 on Intrinsically Disordered Proteins (IDPs). An illustrative example showcasing the AI models' responses to a specific question related to Intrinsically Disordered Proteins (IDPs). This table illustrates the question posed and the corresponding preferred response generated by the GPT-4 model.
Figure 1.
Preferred Response Example from GPT-4 on Intrinsically Disordered Proteins (IDPs). An illustrative example showcasing the AI models' responses to a specific question related to Intrinsically Disordered Proteins (IDPs). This table illustrates the question posed and the corresponding preferred response generated by the GPT-4 model.
Figure 2.
Average Performance Ratings of AI Models Across Different Evaluation Categories. The bar chart presents the average performance ratings of four AI models – GPT-3.5, GPT-4.0, GPT-4.0 with Bing Browser Plugin, and Google Bard (PaLM 2)– across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. Each group of bars corresponds to one of the evaluation categories, and each bar within a group represents the average rating received by a specific AI model in that category. Ratings are on a scale from 1 (lowest) to 5 (highest). The chart provides a comparative assessment of the AI models' performance in handling questions related to Intrinsically Disordered Proteins (IDPs), highlighting their strengths and weaknesses across different aspects of response quality. .
Figure 2.
Average Performance Ratings of AI Models Across Different Evaluation Categories. The bar chart presents the average performance ratings of four AI models – GPT-3.5, GPT-4.0, GPT-4.0 with Bing Browser Plugin, and Google Bard (PaLM 2)– across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. Each group of bars corresponds to one of the evaluation categories, and each bar within a group represents the average rating received by a specific AI model in that category. Ratings are on a scale from 1 (lowest) to 5 (highest). The chart provides a comparative assessment of the AI models' performance in handling questions related to Intrinsically Disordered Proteins (IDPs), highlighting their strengths and weaknesses across different aspects of response quality. .
Figure 3.
Count of AI Models Being Considered as the Best in Handling Questions on Intrinsically Disordered Proteins (IDPs). The bar chart demonstrates the number of times each of the four AI models – GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2)– was considered the best in handling questions related to intrinsically disordered proteins (IDPs). The y-axis represents the count of times an AI model was chosen as the best, and the x-axis lists the AI models. The height of each bar reflects the number of times the corresponding AI model was considered to have provided the most effective response.
Figure 3.
Count of AI Models Being Considered as the Best in Handling Questions on Intrinsically Disordered Proteins (IDPs). The bar chart demonstrates the number of times each of the four AI models – GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2)– was considered the best in handling questions related to intrinsically disordered proteins (IDPs). The y-axis represents the count of times an AI model was chosen as the best, and the x-axis lists the AI models. The height of each bar reflects the number of times the corresponding AI model was considered to have provided the most effective response.
Table 1.
Evaluation Criteria and Rating Scale Utilized in the Expert Evaluation of AI Models. The table outlines the criteria and corresponding 5-point Likert scale used to assess the educational utility of various LLMs within the context of Intrinsically Disordered Proteins (IDPs). The criteria, listed in the left column, include Accuracy, Relevance, Depth of Understanding, Clarity of Explanation, and Quality. The middle column, labeled "Description," provides specific statements that guided the expert evaluator in assigning scores for each criterion. The right column displays the corresponding rating scale, ranging from 1 (indicating strong disagreement or very low quality) to 5 (indicating strong agreement or very high quality). This table exemplifies the systematic evaluation approach adopted to maintain objectivity and consistency in rating the AI models' performance.
Table 1.
Evaluation Criteria and Rating Scale Utilized in the Expert Evaluation of AI Models. The table outlines the criteria and corresponding 5-point Likert scale used to assess the educational utility of various LLMs within the context of Intrinsically Disordered Proteins (IDPs). The criteria, listed in the left column, include Accuracy, Relevance, Depth of Understanding, Clarity of Explanation, and Quality. The middle column, labeled "Description," provides specific statements that guided the expert evaluator in assigning scores for each criterion. The right column displays the corresponding rating scale, ranging from 1 (indicating strong disagreement or very low quality) to 5 (indicating strong agreement or very high quality). This table exemplifies the systematic evaluation approach adopted to maintain objectivity and consistency in rating the AI models' performance.
| Evaluation Criteria |
Description |
| Accuracy |
The information provided in the response is factual and accurate with respect to IDPs. |
| Relevance |
The response directly pertains to the query asked. |
| Depth of Understanding |
The response indicates a profound comprehension of IDPs and related scientific concepts. |
| Clarity of Explanation |
The response is lucid, comprehensible, and well-structured. |
| Quality |
Considering all factors, such as accuracy, relevance, understanding, and clarity, how would you rate the overall quality of the response? |
Table 2.
Assessment Questions and Use Cases for Evaluating AI Models in the Study of Intrinsically Disordered Proteins (IDPs). The table enumerates a series of questions and use cases to evaluate the educational utility of various generative AI models in the context of IDPs. The assessment is aimed at GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Bard, an experimental conversational AI service by Google, powered by PaLM 2. The left column, "Question Set," lists questions probing into the AI models' knowledge and understanding of IDPs. The right column, "Use Cases," provides practical scenarios and recommendations for studying IDPs.
Table 2.
Assessment Questions and Use Cases for Evaluating AI Models in the Study of Intrinsically Disordered Proteins (IDPs). The table enumerates a series of questions and use cases to evaluate the educational utility of various generative AI models in the context of IDPs. The assessment is aimed at GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Bard, an experimental conversational AI service by Google, powered by PaLM 2. The left column, "Question Set," lists questions probing into the AI models' knowledge and understanding of IDPs. The right column, "Use Cases," provides practical scenarios and recommendations for studying IDPs.
| Question Set |
Use Cases |
| How has the traditional “one structure, one function” paradigm limited our understanding of protein functionality and roles of proteins in biological systems? |
Recommend an experimental procedure for characterizing the folding landscape and structural transitions of an intrinsically disordered protein using biophysical techniques. |
How do intrinsically disordered proteins challenge the structure-function paradigm, and what evidence supports their importance in cellular processes?
|
Propose a computational approach to simulate the conformational behavior and dynamics of intrinsically disordered proteins. |
What are some misconceptions about intrinsically disordered proteins that persist in the scientific community, and how can they be addressed?
|
Design an experimental protocol to demonstrate the involvement of intrinsically disordered proteins in liquid-liquid phase separation. |
What are the most significant findings or breakthroughs in the study of intrinsically disordered proteins that have changed our understanding of protein structure and function?
|
Develop a pipeline for integrating multiple experimental and bioinformatics datasets to identify novel intrinsically disordered protein candidates and prioritize their functional characterization. |
| How can the study of intrinsically disordered proteins contribute to a more comprehensive understanding of protein-protein interactions and cellular signaling networks? |
Outline a research plan to elucidate the molecular mechanisms underlying the aggregation and pathological behavior of disease-associated intrinsically disordered proteins, considering both in vitro and cellular models. |
What are the roles of intrinsically disordered proteins in liquid-liquid phase separation and biogenesis of membrane-less organelles?
|
|
How can the development of novel experimental techniques specifically tailored to study intrinsically disordered proteins advance our understanding of their structure, function, and interactions?
|
|
How can incorporating the study of intrinsically disordered proteins into drug discovery efforts lead to the identification of novel therapeutic targets and strategies?
|
|
How can we improve the acceptance and understanding of intrinsically disordered proteins in the scientific community and promote their importance in the biological sciences?
|
|
| What are the future directions and challenges in the field of intrinsically disordered proteins research, and how can the scientific community work together to address these challenges? |
|
Table 3.
Descriptive Statistics of AI Models’ Performance Acoss Evaluation Categories. The table presents the descriptive statistics, including mean and standard deviation, of the performance ratings received by the AI models, GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2) - across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. The rows represent the AI models, while the columns represent the evaluation categories, subdivided into mean and standard deviation.
Table 3.
Descriptive Statistics of AI Models’ Performance Acoss Evaluation Categories. The table presents the descriptive statistics, including mean and standard deviation, of the performance ratings received by the AI models, GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2) - across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. The rows represent the AI models, while the columns represent the evaluation categories, subdivided into mean and standard deviation.
| |
Accuracy |
Relevance |
Understanding |
Clarity |
Quality |
| Model |
mean |
std |
mean |
std |
mean |
std |
mean |
std |
mean |
std |
| GPT-3.5 |
4.87 |
0.35 |
4.93 |
0.26 |
4.67 |
0.62 |
5.00 |
0.00 |
4.87 |
0.35 |
| GPT-4 |
4.93 |
0.26 |
5.00 |
0.00 |
4.93 |
0.26 |
5.00 |
0.00 |
4.93 |
0.26 |
| GPT-4 with Browsing |
4.07 |
0.46 |
4.47 |
0.64 |
3.87 |
0.64 |
4.53 |
0.52 |
4.07 |
0.46 |
| Google Bard (PaLM 2) |
4.20 |
0.56 |
4.47 |
0.52 |
3.87 |
0.52 |
4.80 |
0.41 |
4.13 |
0.35 |
Table 4.
Results of Tukey's Honestly Significant Difference (HSD) Test for AI Model Performance Across Evaluation Categories. The table presents the results of Tukey's Honestly Significant Difference (HSD) test, which was conducted to assess pairwise differences between the AI models – GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2) – across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. For each pairwise comparison, the table shows the mean difference (meandiff), the confidence interval, and the decision to reject or not reject the null hypothesis of no difference between the groups. For additional visualizations, statistically significant differences are shaded.
Table 4.
Results of Tukey's Honestly Significant Difference (HSD) Test for AI Model Performance Across Evaluation Categories. The table presents the results of Tukey's Honestly Significant Difference (HSD) test, which was conducted to assess pairwise differences between the AI models – GPT-3.5, GPT-4, GPT-4 with Bing Browser Plugin, and Google Bard (PaLM 2) – across five evaluation categories: accuracy, relevance, depth of understanding, clarity of explanation, and overall quality. For each pairwise comparison, the table shows the mean difference (meandiff), the confidence interval, and the decision to reject or not reject the null hypothesis of no difference between the groups. For additional visualizations, statistically significant differences are shaded.
| Tukey's HSD Test Results for Accuracy |
| Group 1 |
Group 2 |
meandiff |
p-adj |
lower |
upper |
reject |
| GPT-3.5 |
GPT-4 |
0.0667 |
0.9 |
-0.3419 |
0.4753 |
False |
| GPT-3.5 |
GPT-4 with Browsing |
-0.8 |
0.001 |
-1.2086 |
-0.3914 |
True |
| GPT-3.5 |
Google Bard (PaLM 2) |
-0.6667 |
0.001 |
-1.0753 |
-0.2581 |
True |
| GPT-4 |
GPT-4 with Browsing |
-0.8667 |
0.001 |
-1.2753 |
-0.4581 |
True |
| GPT-4 |
Google Bard (PaLM 2) |
-0.7333 |
0.001 |
-1.1419 |
-0.3247 |
True |
| GPT-4 with Browsing |
Google Bard (PaLM 2) |
0.1333 |
0.8011 |
-0.2753 |
0.5419 |
False |
| Tukey's HSD Test Results for Relevance |
| Group 1 |
Group 2 |
meandiff |
p-adj |
lower |
upper |
reject |
| GPT-3.5 |
GPT-4 |
0.0667 |
0.9 |
-0.35 |
0.4834 |
False |
| GPT-3.5 |
GPT-4 with Browsing |
-0.4667 |
0.0224 |
-0.8834 |
-0.05 |
True |
| GPT-3.5 |
Google Bard (PaLM 2) |
-0.4667 |
0.0224 |
-0.8834 |
-0.05 |
True |
| GPT-4 |
GPT-4 with Browsing |
-0.5333 |
0.0069 |
-0.95 |
-0.1166 |
True |
| GPT-4 |
Google Bard (PaLM 2) |
-0.5333 |
0.0069 |
-0.95 |
-0.1166 |
True |
| GPT-4 with Browsing |
Google Bard (PaLM 2) |
0 |
0.9 |
-0.4167 |
0.4167 |
False |
| Tukey's HSD Test Results for Understanding |
| Group 1 |
Group 2 |
meandiff |
p-adj |
lower |
upper |
reject |
| GPT-3.5 |
GPT-4 |
0.2667 |
0.5165 |
-0.2458 |
0.7792 |
False |
| GPT-3.5 |
GPT-4 with Browsing |
-0.8 |
0.001 |
-1.3125 |
-0.2875 |
True |
| GPT-3.5 |
Google Bard (PaLM 2) |
-0.8 |
0.001 |
-1.3125 |
-0.2875 |
True |
| GPT-4 |
GPT-4 with Browsing |
-1.0667 |
0.001 |
-1.5792 |
-0.5542 |
True |
| GPT-4 |
Google Bard (PaLM 2) |
-1.0667 |
0.001 |
-1.5792 |
-0.5542 |
True |
| GPT-4 with Browsing |
Google Bard (PaLM 2) |
0 |
0.9 |
-0.5125 |
0.5125 |
False |
| Tukey's HSD Test Results for Clarity |
| Group 1 |
Group 2 |
meandiff |
p-adj |
lower |
upper |
reject |
| GPT-3.5 |
GPT-4 |
0 |
0.9 |
-0.32 |
0.32 |
False |
| GPT-3.5 |
GPT-4 with Browsing |
-0.4667 |
0.0016 |
-0.7867 |
-0.1467 |
True |
| GPT-3.5 |
Google Bard (PaLM 2) |
-0.2 |
0.3575 |
-0.52 |
0.12 |
False |
| GPT-4 |
GPT-4 with Browsing |
-0.4667 |
0.0016 |
-0.7867 |
-0.1467 |
True |
| GPT-4 |
Google Bard (PaLM 2) |
-0.2 |
0.3575 |
-0.52 |
0.12 |
False |
| GPT-4 with Browsing |
Google Bard (PaLM 2) |
0.2667 |
0.1338 |
-0.0533 |
0.5867 |
False |
| Tukey's HSD Test Results for Quality |
| Group 1 |
Group 2 |
meandiff |
p-adj |
lower |
upper |
reject |
| GPT-3.5 |
GPT-4 |
0.0667 |
0.9 |
-0.2832 |
0.4166 |
False |
| GPT-3.5 |
GPT-4 with Browsing |
-0.8 |
0.001 |
-1.1499 |
-0.4501 |
True |
| GPT-3.5 |
Google Bard (PaLM 2) |
-0.7333 |
0.001 |
-1.0832 |
-0.3834 |
True |
| GPT-4 |
GPT-4 with Browsing |
-0.8667 |
0.001 |
-1.2166 |
-0.5168 |
True |
| GPT-4 |
Google Bard (PaLM 2) |
-0.8 |
0.001 |
-1.1499 |
-0.4501 |
True |
| GPT-4 with Browsing |
Google Bard (PaLM 2) |
0.0667 |
0.9 |
-0.2832 |
0.4166 |
False |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).