1. Introduction
Triatoma infestans (Klug) is a hemipteran insect belonging to the Reduviidae family and the most important vector of Chagas disease. This species developed through the Andean and non-Andean evolutionary lineages with different geographic distributions in South America, genomic size, and main repetitive DNA [
1]. Both lineages dispersed from Bolivian highlands, from where they had originated. Forty-two satellite DNA families are present in varying amounts in both lineages [
2].
The Andean lineage was primarily dispersed in the high-altitude valleys of Bolivia and Peru and in lower altitudes of southern Peru; the non-Andean lineage occupied lowland regions in Argentina, Brazil, Chile, Paraguay, Uruguay, and Bolivia and certain higher altitude regions in Argentina until chemical interventions for insect control were undertaken in the past decades, which drastically reduced these groups [
2]. Although these lineages differ in nuclear DNA content per haploid genome (1,487 Gbp, – non-Andean lineage; 1,936 Gbp, – Andean lineage) and in the amounts of satellite DNA families, they have the same number of holokinetic chromosomes (2n = 20 + XY or XX) [
2].
In addition to the importance of T. infestans as a vector for Trypanosoma cruzi infection, it has been revealed as a model for studies on unusual cellular processes involving somatic polyploidy, nuclear and cell fusion, presence of large heterochromatin bodies (chromocenters), diversity of nuclear phenotypes, cell death, epigenetic markers, and chromatin response to histone deacetylase inhibition. This unprecedented review briefly describes these phenomena in T. infestans specimens collected in Brazil and subsequently reared in insectary facilities at 30 °C and 80% relative humidity. The somatic cell peculiarities reported here point out T. infestans as an attractive model for cell biology investigations.
2. Somatic Polyploidy
Insects of most Orders develop somatic polyploidy, in which DNA endoreplication steps advance and permit nuclear and cellular volume enlargement and organ growth without intervention of mitosis [
3]. This phenomenon occurs mainly during the larval stages in insects undergoing complete metamorphosis (holometabola), or in nymphal instars of insects undergoing simple metamorphosis (hemimetabola) [
3]. Hemipterans develop through hemimetaboly. Somatic polyploidy involving an integral doubling of the genome does not occur in all insect groups; specific genomic regions as in the case of the heterochromatin may replicate differentially [
4].
Somatic polyploidy may be considered an adaptive process through which the cells save the physiological time and protein synthesis that would be required if a mitotic spindle and nuclear membrane were fabricated as in the case of ordinary somatic division [
3]. Some authors have hypothesized that it is a strategy for increasing gene copy numbers, which would facilitate high rates of biosynthesis and metabolism [
4]. In healthy mammal tissues, somatic polyploidy has been considered important for cells to function under stress, protecting their viability and permitting their adaptation under new environmental conditions [
5,
6].
The DNA endoreplication steps that occur in the nuclei of the Malpighian tubule cells of fully-nourished
T. infestans specimens are arrested at the end of the 5th nymphal instar, when 32 C and 64 C optimal ploidy degrees are attained per nucleus compared with the DNA content of nuclei in the first nymphal stages and in spermatids [
7,
8]. However, in fasted insects, the ordinary polyploidization pattern detected in the Malpighian tubules is surpassed because nuclear and cellular fusions occur even with low incidence in specimens reared in the laboratory [
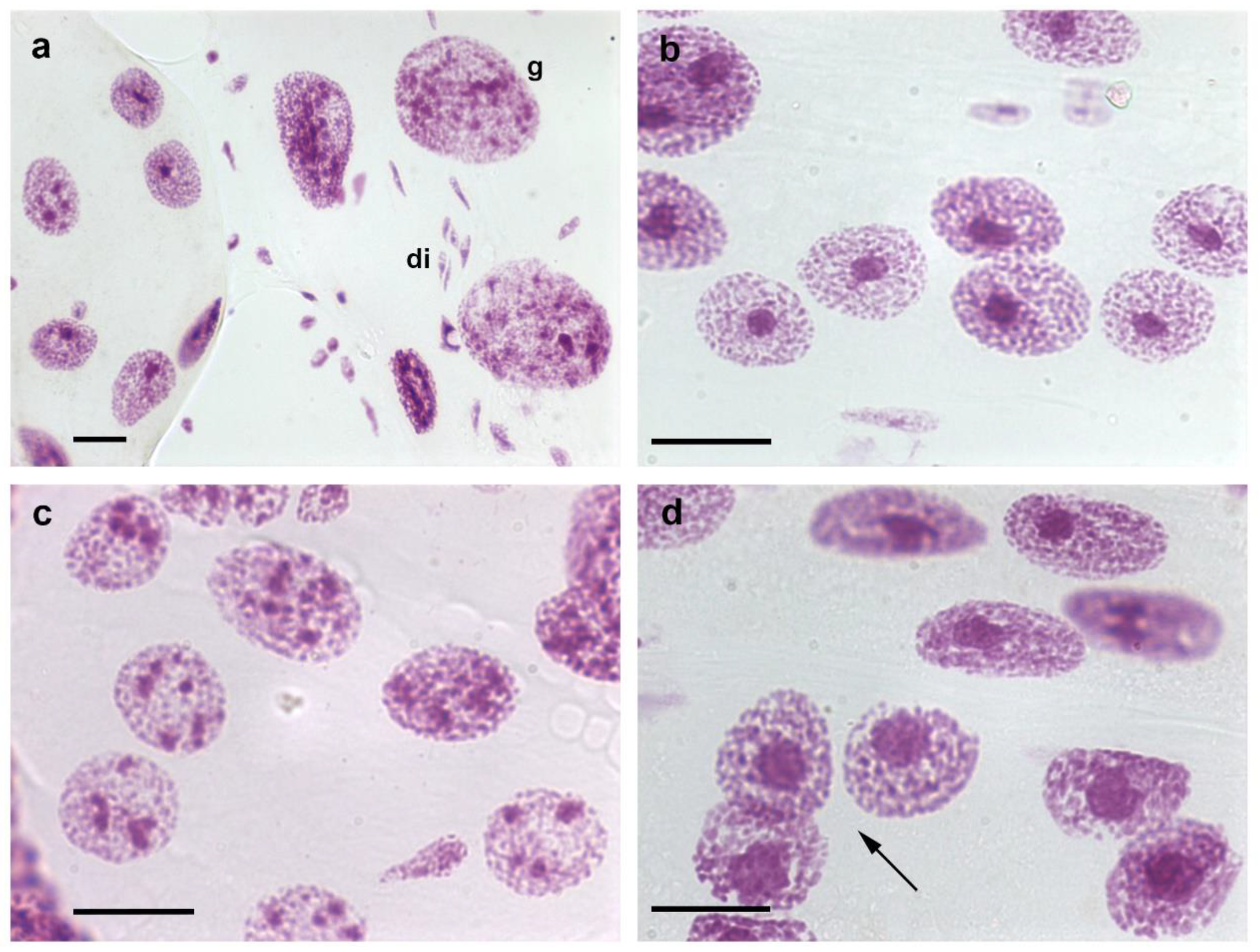
9]. Then, ploidy degrees shift to considerably higher values, as demonstrated by DNA cytophotometric quantification, visual detection of the presence of giant nuclei (
Figure 1a), or counting of the total number of nuclei/organ that decreases significantly [
10]. Giant nuclei may attain a ploidy degree as high as 1024 C [
10]. Consequently, the total number of nuclei may show a 40% reduction [
11]. Additionally, nuclear and cell fusions have been detected in BTC-32 cells cultivated from embryonic tissues of
T. infestans at the London School of Hygiene and Tropical Medicine [
12]. The presence of giant nuclei is not restricted to
T. infestans; it has also been reported in the fat body cells of
Rhodnius prolixus, another species of the Reduviidae family [
13].
Initially, nuclear fusion is facilitated by the binucleate composition of the somatic cells [
10]. When fasting periods are prolonged, cell fusion and additional nuclear fusions occur in the Malpighian tubules. Cell fusion may be favored by the presence of the viral particles that infect these cells, especially when the insects are fed avian blood under laboratory conditions [
14,
15]. Cell fusion has been suggested to represent an attempt at cell survival that opposes cell death [
10,
13]; however, both phenomena may coexist in the same organ [
9].
3. Heterochromatin and Nuclear Phenotypes
Condensed chromatin aggregates comprising constitutive heterochromatin (chromocenters) are present in
T. infestans cells from the 1st nymphal instar [
7]. These bodies increase in size through DNA endoreplication steps that do not always accompany the DNA doubling process in the euchromatin (DNA underreplication), representing a constitutive heterochromatin characteristic [
7]. Other indicators of the constitutive heterochromatin nature of these condensed chromatin bodies are the absence of RNA transcriptional activity in most regions of these bodies, as detected using radioautography; abundance of repetitive DNA; and differential response to the Feulgen reaction compared with the characteristics of euchromatin [
7,
16,
17].
The chromocenters of
T. infestans are usually composed of copies of the large A, B, and C autosomes plus the X and Y sex chromosomes [
18,
19]. One additional autosome pair has been reported as part of this heterochromatin type under specific insect rearing conditions [
20]. Additionally, polymorphism and variable amount of heterochromatin have been reported for
T. infestans [
21,
22,
23].
From the 1st to the 3rd nymphal instar, only one chromocenter is present in the Malpighian tubules of
T. infestans; from this stage onward, a portion of the cells exhibit more than one chromocenter (
Figure 1b,c) [
7,
8]. The process of formation of multi-chromocenters apparently involves budding or de-clustering from the single chromocenter body [
24] and does not lead to the micronuclei production that results in cell death as reported for other cell systems [
25]. The nuclear phenotype diversification and the size attained by chromocenters in the Malpighian tubules of nymphs and adults have not been identified in other species of the Reduviidae family studied till date [
26]. These unexplained phenomena could not be shown to be associated with diversified functions exerted by the Malpighian tubules, which are excretory organs eliminating the byproducts of a blood diet that does not change during the insect lifetime.
Notably, free DNA phosphates showed differential availability for the electrostatic binding of toluidine blue molecules under Mg
2+ competitive staining conditions, when the single-chromocentered nuclei were compared with nuclei bearing multiple chromocenters [
27]. This finding revealed that the dye-binding sites in the DNA of chromocenters in the multi-chromocentered nuclei are more exposed and easier to block by Mg
2+ ions than those in the DNA of chromocenters in single-chromocentered nuclei probably owing to differences in the packing states of the DNA-protein complexes involved [
27].
T. infestans nuclei constitute an attractive model for studying the 3D position of chromocenters because of the large volume of these bodies inside the nuclei. Using confocal microscopy and quantitative measurement of the 3D position of the chromocenters in the Malpighian tubule cells of
T. infestans, the proximity of these bodies to the nuclear periphery was demonstrated during the insect lifetime [
24]. The spatial distribution of chromocenters in relation to the nuclear periphery was confirmed using electron microscopy [
24].
The proximity of chromocenters to the nuclear periphery has been detected in the cells of several other organisms [
28,
29,
30,
31], including species of the Reduviidae family which exhibit very small chromocenters, as in the case of
Panstrongylus megistus [
32]; however, the reason for this phenomenon remains unclear [
33]. The concept that proximity of a chromocenter to the nuclear periphery is associated with overall gene silencing [
34,
35] does not fully apply to
T. infestans, because active 18 S rDNA sites were detected in the chromocenter surface including the region facing the nuclear periphery, according to videos obtained after 3D fluorescence
in situ hybridization [
24]. In
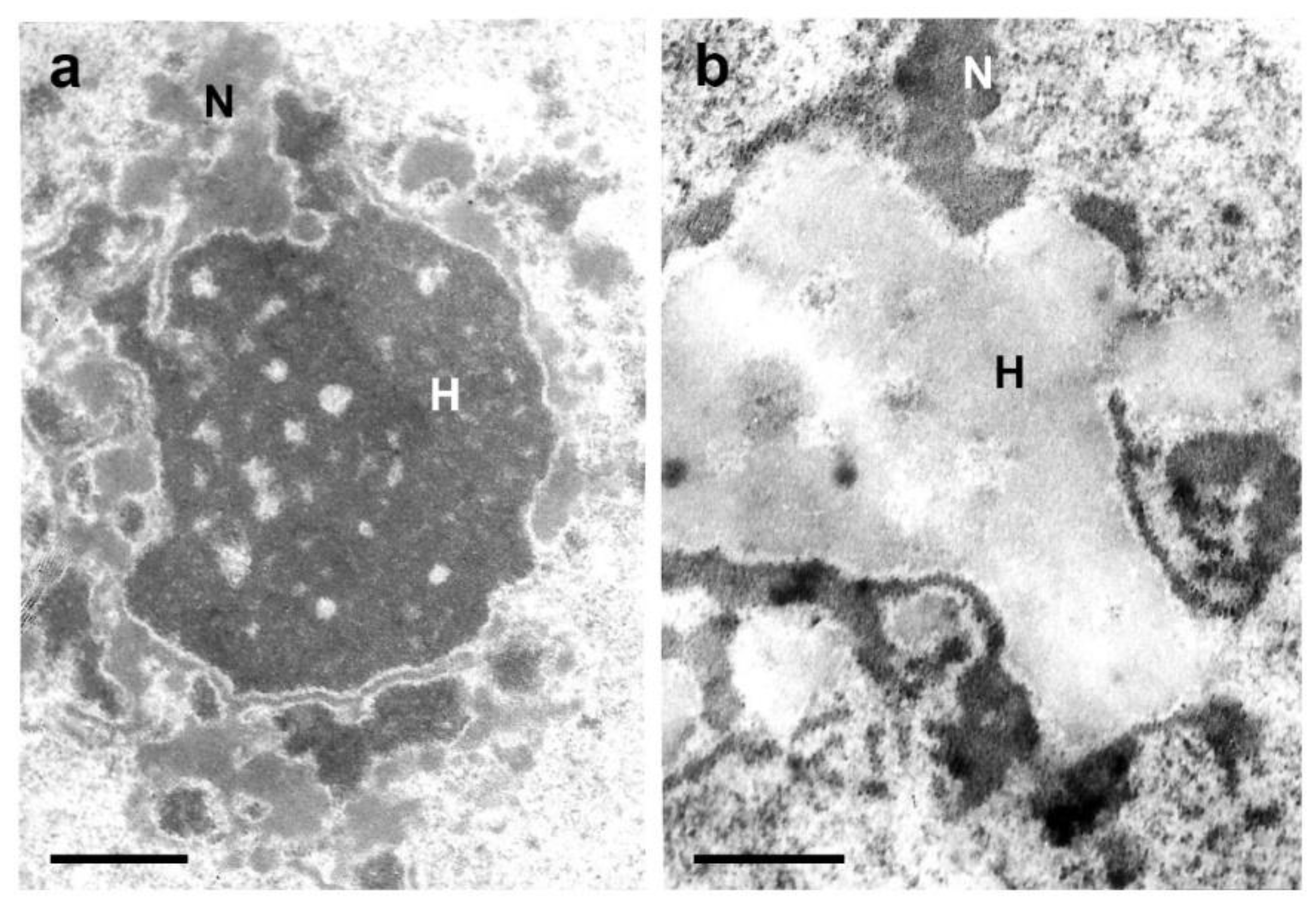
T. infestans cells, the nucleolus was found to encircle chromocenters (
Figure 2a,b) [
36]. Additionally, the hypothesis that heterochromatin as a gene-silencing compartment has been questioned [
29]. Variability relative to the small distance between the edge of the chromocenters and the nuclear periphery raises the suspicion of the mobility of the heterochromatin body with respect to the nuclear envelope [
24].
Unraveling of the chromocenter heterochromatin has been observed in a small number of nuclei in the Malpighian tubules of
T. infestans under rearing conditions in the laboratory (
Figure 1d) [
9]. However, the frequency of this phenotype was observed to be significantly increased in fasted nymphs and in specimens subjected to heat shock [
9,
10]. This phenomenon, which affects the heterochromatin in other cell systems and under different physiological conditions has been hypothesized to represent an attempt to activate silent genes possibly present in this chromatin compartment and achieve cell survival [
37,
38].
Jagannathan et al. [
25,
39], through studies in
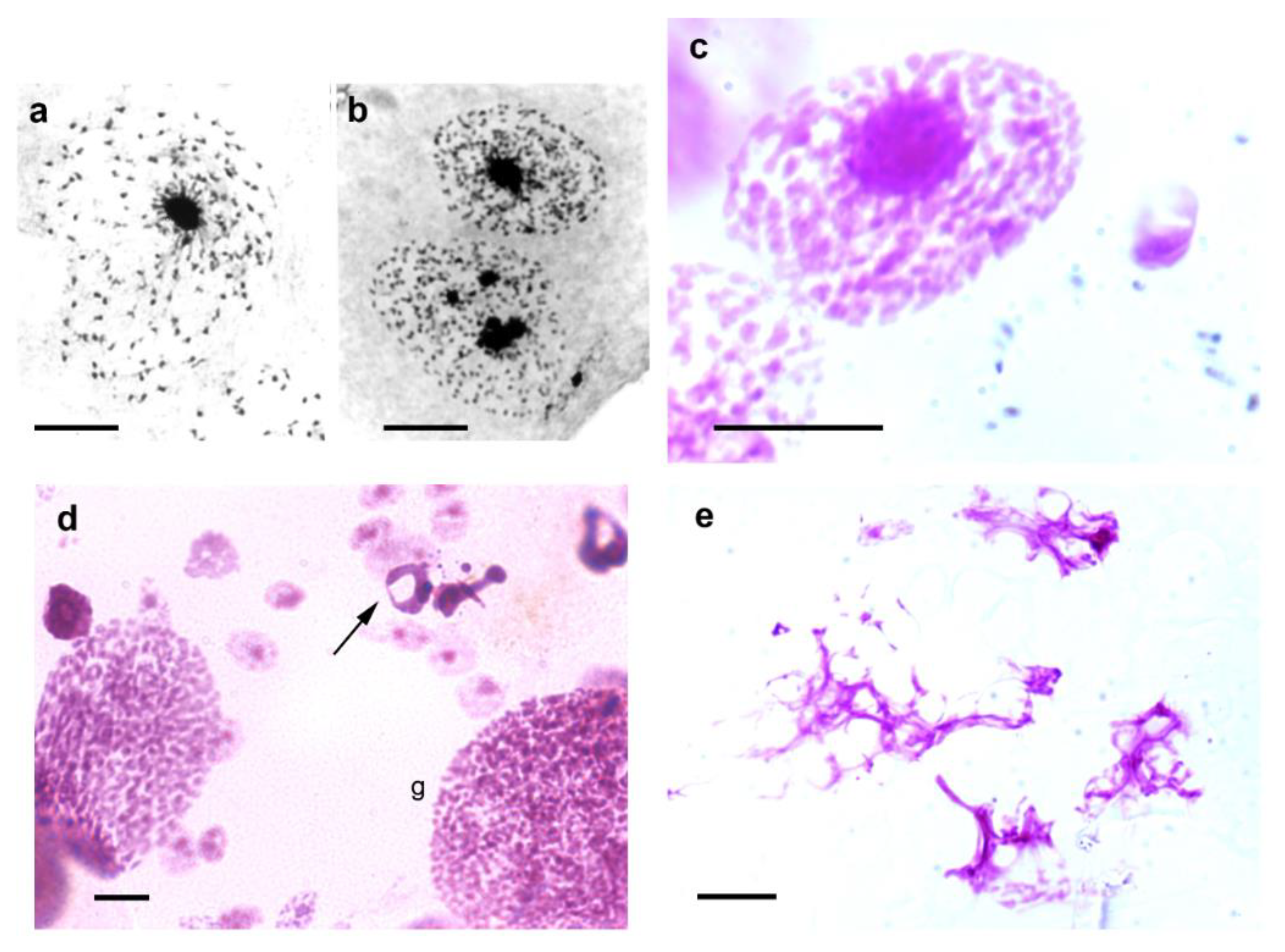
Drosophila and mouse cells, proposed that satellite DNA and their binding proteins may physically integrate the entire chromosome complement into a chromocenter network thus ensuring encapsulation of all chromosomes within a single cell. Notably, images of the epithelial cell nuclei of
T. infestans suggest confluence of chromosomes that do not properly integrate the chromocenters into this heterochromatin cluster (
Figure 3a–c).
4. Response to Stress Agents
T. infestans is resistant to long fasting periods. The Malpighian tubules of 5th instar nymphs starved for 6.5 months not only exhibit an increased frequency of giant nuclei, while the frequency of single- and multi-chromocentered nuclei decreases, but also an increased frequency of the nuclear phenotype in which chromocenter decondensation is evident [
10]. In addition to the nuclear phenotypes usually detected in the Malpighian tubules of fully-nourished specimens of
T. infestans, and the giant nuclei resulting from cell fusion, nuclei with morphological characteristics representing cell death or exhibiting chromocenter decondensation may be detected under stress conditions (
Figure 3d,e).
Heat and cold shocks have been reported to elicit different responses with respect to survival of the specimens and frequency of nuclear phenotypes in the Malpighian tubules compared with fasting conditions. Seven days after a 1-hour-heat shock treatment at 40 °C, the frequency of single- and multi-chromocentered nuclei decreased while the frequency of the nuclear phenotype with chromocenter decondensation increased, giant nuclei were few, and nuclear images showed cell death types (apoptosis and especially necrosis) [
9]. Thirty days after the 1-h-heat shock treatment in fasted specimens, the frequency of giant nuclei increased, while that of nuclei exhibiting chromocenter decondensation was low [
9]. Subsequently, giant nuclei affected by apoptosis [
40], as well as necrosis substituting for apoptosis, were also observed [
9]. Whether the presence of giant nuclei and nuclei with chromocenter unpacking can be considered cellular survival attempts, they were not effective enough to cope with the metabolic events induced by the 1-hour heat-shock treatment, because the cell death effects surpassed the survival attempts.
When insects were subjected to heat shock at 40 °C for 12 h, the frequency of the Malpighian tubule nuclei containing unpacked chromocenters reached up to 33% relative to the total nuclei immediately after the shock, and reduces abruptly to 3.35% and 0.39% 3 and 30 days, respectively, following the shock [
41]. Conversely, the frequency of the giant nuclei increased up to 8.5% relative to the total nuclei in the 30-day period. Although the frequency of cell death images did not surpass 3.5% of the cell nuclei in the Malpighian tubules from insects 30 days after the 12-h heat shock, the total number of nuclei of approximately 12,000 in the controls reduced to 4,500-6,000 after this shock [
41].
In the Malpighian tubules of
T. infestans, giant nuclei have been reported to be few in nymphs examined 30 days after a 0 °C-shock for 1 h and unobservable after a cold shock for 12 h. Chromocenter unpacking is rare under both cold-shock periods. While apoptosis is also rare, necrosis frequency may reach approximately 7% relative to the total cell nuclei 30 days after the 12-h cold shock [
41].
Regarding survival of the
T. infestans specimens subjected to temperature shocks, their resistance to heat and cold shock for 1 h at 40 °C and 0 °C has been demonstrated to some extent, despite the above-mentioned cellular effects visualized in their Malpighian tubules. Heat and cold shock conditions did not affect nymphal survival in a 30-day period, although they affected the hormonal balance which controls molting and slightly decreased the survival of male and female adults [
42]. However, when heat shock periods at 40 °C were extended to 12 h, survival decreased to 28% in 3rd instar nymphs, 32% in male adults, and 52% in female adults. Nevertheless, the 4th and 5th instar nymphs of
T. infestans were not found to be affected by this long shock [
42].
Fully-nourished or fasted fifth instar specimens of
T. infestans have shown tolerance to cold shock at 0 °C for up to 12 h even if subjected to a single shock or to sequential shocks at intervals of 8 or 24 h at 30 °C and examined 32 days later [
43]. This response to cold shock may have been enabled the species to survive low temperatures at their original altitudes in the Andes Mountains before adaptation to Neotropical areas throughout regions of the South America. Such a survival response differs significantly from the less than 10% survival of fifth instar nymphs of
Panstrongylus megistus after a 12 h shock at 0 °C [
43,
44].
P. megistus is another reduviid species vector of Chagas disease and well disseminated in Brazil.
T. infestans showed a decreased frequency of apoptosis and necrosis in the Malpighian tubules following sequential cold shocks under full nourishment or 15-day fasting before the initial shock, confirming the tolerance of this insect to cold stress [
45].
In a study on the response of
T. infestans to heavy metal ions, another stress agent, fasted 5th nymphal specimens showed 90% survival up to 24 h after injection of 2 x 10
-5 M CuSO
4 and 1.56 x 10
-4 M CH
3HgCl; however, after this period, survival abruptly decreased, especially in fully-nourished specimens. In the presence of these heavy metals, necrosis was the most frequent phenotype in the Malpighian tubules, occurring predominantly 2 h after treatments; less than 20% of the single- and multi-chromocentered nuclei exhibited unpacked chromocenters, and very few giant nuclei and apoptotic nuclei were detected [
46]. These findings demonstrated the lack of resistance of
T. infestans to heavy metals.
5. Epigenetic Markers: DNA and Histone Methylation
The chromocenters of both single- and multi-chromocentered nuclei of
T. infestans contain AT-rich, GC-poor DNA, as demonstrated using cytochemistry and immunofluorescence analysis [
17]. Cytogenetics, immunolabeling, and molecular biology assays have identified at least four chromosome rearrangements involved in the amplification/dispersion of AT-rich satellite DNA [
23]. Fourier transform infrared (FTIR) microspectroscopy confirmed AT-rich DNA in the whole nuclear genome of
T. infestans [
47].
While DNA 5-methylcytosine (5mC) has not been immunocytochemically revealed in chromocenters of the Malpighian tubules of
T. infestans, it occurs in the euchromatin that is especially concentrated around the chromocenters [
17,
47]. This absence of 5mC in the heterochromatin is not limited to
T. infestans; it has also been reported in hemipteran coccids and aphid species [
48,
49].
Histone epigenetic markers such as acetylated H3K9 (H3K9ac) and H4K8 (H4K8ac) and methylated and di-methylated H3K9 (H3K9me/me2) and H4K20 (H4K20me/me2) have not been detected immunocytochemically in the chromocenters of both single- and multi-chromocentered nuclei of
T. infestans. However, they have been detected in the euchromatin of these nuclei [
50]. These findings are consistent with the differential identification of such markers, except H3K9me2, in the euchromatin compared with that in the heterochromatin in other cell systems [
51,
52,
53,
54]. H3K9me2 is generally associated with gene silencing and constitutive heterochromatin formation [
51,
55]. However, the chromocenters of
T. infestans do not exhibit positive fluorescence signals in response to anti-H3K9me2 antibody, while euchromatin is positively stained [
50]. H4K8ac and H3K9me signals in euchromatin appear concentrated around the chromocenter in single-chromocentered nuclei [
50]. This is the region where active 18-S rDNA sites have been demonstrated [
24].
The immunofluorescence signals for acetylated H4K16 (H4K16ac) in the images reported for
T. infestans are not sufficiently clear to discard the presence of this marker in the heterochromatin body [
50]. In addition, H4K16ac has been associated with gene transcriptional activities by some authors [
56,
57] but with transcriptional repression by others [
58]. Therefore, the results reported for this marker in
T. infestans and their implications are inconclusive till date and require in-depth investigation possibly using 3D confocal microscopy and molecular biology assays.
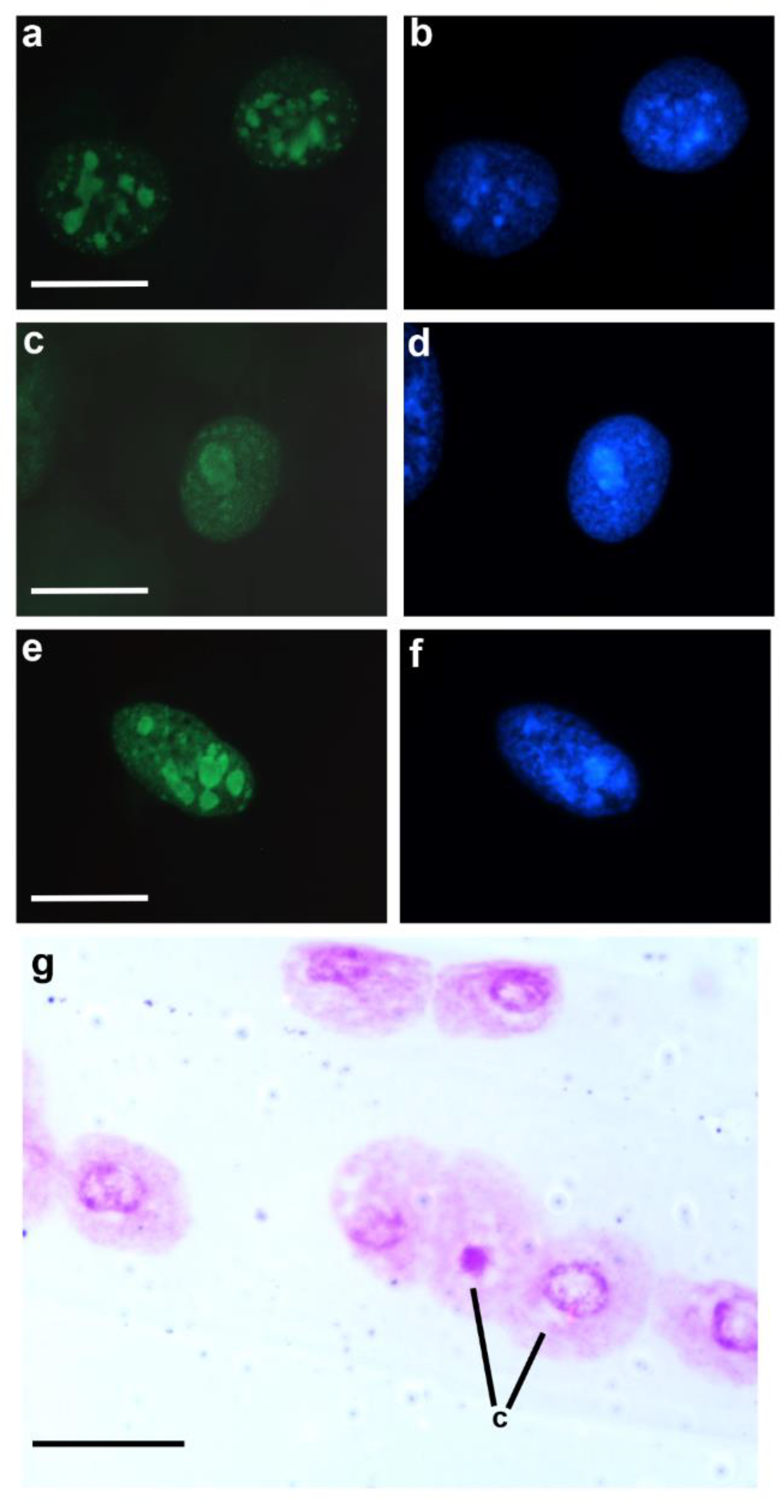
Tri-methylated H3K9 (H3K9me3) marker predominates in the chromocenters of single- and multi-chromocentered nuclei of
T. infestans; additionally, positive signals are observed in euchromatin granular points (
Figure 4a,b) [
50]. This epigenetic marker along with methylated H3K27 has been associated with constitutive heterochromatin and inactive enhancers in other cell systems [
54,
59]. The finding of H3K9me3 signals in euchromatin points may indicate their position in promoters of repressed genes [
50]. Tri-methylated H4K20 (H4K20me3), that is considered a constitutive heterochromatin hallmark [
60,
61], is also revealed in the chromocenters of
T. infestans (
Figure 4c–f).
6. Response to Epigenetic Modulators
Valproic acid (VPA) is a short-chain branched fatty acid, and its application extends from beyond its primary and wide use in the treatment of neurological disorders to the modulation of epigenetic markers, including inhibition of histone deacetylases, which favors histone acetylation; changes in the DNA and histone methylation status; direct binding to chromatin components; and chromatin remodeling [
54,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72]. VPA has also been revealed as a promising agent against a series of tumors, owing to its inhibitory effects on cell proliferation and induction of cell cycle arrest and cell death [
73,
74,
75,
76].
Insects have been suggested as an alternative model for research on epigenetic effects promoted by VPA, because of their shorter generation intervals, the opportunity for analyzing larger samples, and diminished ethical concerns compared with those for the experimental use of mammals. Therefore,
T. infestans may be considered an attractive candidate for evaluating pathway mechanisms underlying VPA action [
77].
In the presence of VPA,
T. infestans cells revealed unexpected results [
50,
77,
78]. One-hour following the abdominal injection of a 0.05 mM VPA solution in
T. infestans nymphs or after a 4-h
in vitro culture of the Malpighian tubules of this insect in the presence of 0.05-0.5 mM VPA, unpacking of the chromocenter was demonstrated in a portion of the Malpighian tubule cells (
Figure 4g) [
50,
78]. Higher VPA concentrations lead to co-occurrence of apoptosis and necrosis in the cells of this tissue [
78]. Observation of chromocenter unpackaging, which was not accompanied by induction of H3K9 acetylation, differed from the reports for other cell types [
67]. An alternative explanation for this finding would be that DNA cytosine demethylation occurred following VPA treatment. However, this hypothesis does not apply to
T. infestans chromocenters because their DNA is poor or lacking in 5mC [
47]. No explanation has been proposed for the induction of the chromocenter decondensation in
T. infestans cells in the presence of VPA, not involving H3K9 acetylation and DNA demethylation. It certainly requires investigation of other molecules participating in the orchestration of the condensed state of the chromocenters, such as H3K9me3, H4K20me3, the heterochromatin-associated protein (HP1-α) and other non-histone proteins. In other cell systems, H3K9me3 furnishes a binding site to HP1 [
79,
80]. Regulation of H4K20me3 can occur via an interaction between methyltransferase KMT5C and HP1 [
60].
7. Concluding Remarks
In addition to its relevance as a vector of a disease that affects more than 6 to 7 million people in the Americas,
T. infestans calls attention as a model for certain cellular peculiarities that may have evolved and contributed to its biological resilience. Most unanswered questions rely on the nature and role of the chromocenters in the Malpighian tubules of this insect. Regardless of whether changes in the number of chromocenters per nucleus are associated with changes in cell fate [
31], the disruption of the single chromocenter affecting only a portion of the nuclei in the Malpighian tubules after the 3rd nymphal instar remains to be explained. If chromocenter formation contributes to nuclear stiffening and resistance to small nuclear deformations independent of lamins [
81], the de-clustering of a single chromocenter into a variable number of chromocenters, with no loss of structural and mechanical nuclear integrity, requires interpretation. The hypothetical role of chromocenters in forming a network with chromosomes that do not properly integrate the heterochromatin body needs demonstration of involvement of binding proteins to satellite DNA as reported for mouse and
Drosophila chromocenters [
25,
39]. Mobility of the chromocenters inside the cell nuclei, as assumed based on the variability of the distance between the chromocenters and the nuclear periphery, could be supported by
in vivo observations. Further studies are also necessary to determine the implication of unraveling of the chromatin encapsulated into chromocenter bodies under stress conditions or in response to the epidrug VPA, with no association of H3K9 acetylation. These studies include examination and analysis of 3D images representative of other epigenetic markers. Further research on these questions would certainly unveil new and exciting findings that corroborate the use of
T. infestans as a unique biological model.
Author Contributions
M.L.S.M. conceived and wrote the original draft of the manuscript, and has read and agreed to the published version of the manuscript.
Funding
This investigation was supported by grants from the São Paulo State Research Foundation (FAPESP, Brazil; grant no. 2015/10356-2) and Brazilian National Council for Research and Development (CNPq, grant no.304797/2019-7). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Informed Consent Statement
Not applicable.
Acknowledgments
The author is indebted to Dr. Douglas Santos for his assistance with the immunocytochemistry for H4K20me3, to Mr. Eli H.M. dos Anjos for formatting the micrographs, and to Prof. Dr. Benedicto de Campos Vidal for critically reading the manuscript.
Conflicts of Interest
The author declares no conflict of interest.
References
- Bargues, M.D.; Kiisiowicz, D.R.; Panzera, F.; Noireau, F.; Marcilla, A.; Perez, R.; Rojas, M.G.; O’Connor, J.E.; Gonzales-Candelas, F.; Galvao, C.; Jurberg, J.; Carcavallo, R.U.; Dujarding, J.; Mas-Coma, S. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Infect. Genet. Evol. 2006, 6, 436–462. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Mora, P.; Vel, J.; Cuadrado, A.; Sanchez, A.; Palomeque, T.; Lorite, P. Comparative repeatome analysis on Triatoma infestans Andean and Non-Andean lineages, main vector of Chagas disease. PLoS ONE 2017, 12, e0181635. [Google Scholar] [CrossRef]
- Mello, M.L.S. Somatic polyploidy in insects. Ciênc. Cult. 1970, 22, 348–350. [Google Scholar]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Somatic polyploidy promotes cell function under stress and energy depletion: evidence from tissue-specific mammal transcriptome. Funct. Integr. Genomics 2010, 10, 433–446. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy as a fundamental phenomenon in Evolution, Development, Adaptation and Diseases. Int. J. Mol. Sci. 2022, 23, 3542. [Google Scholar] [CrossRef]
- Mello, M.L.S. Nuclear behavior in the Malpighian tubes of Triatoma infestans (Reduv., Hemiptera). Cytologia 1971, 36, 42–49. [Google Scholar] [CrossRef]
- Mello, M.L.S. Feulgen-DNA values and ploidy degrees in the Malpighian tubes of some triatomids. Revta. Brasil. Pesq. Méd. Biol. 1975, 8, 101–107. [Google Scholar]
- Mello, M.L.S.; Tavares, M.C.H.; Dantas, M.M.; Rodrigues, V.L.C.C.; Maria-Engler, S.S.; Campos, S.P.; Garcia, N.L. Cell death and survival alterations in Malpighian tubules of Triatoma infestans following heat shock. Biochem. Cell Biol. 2001, 79, 709–717. [Google Scholar] [CrossRef]
- Mello, M.L.S. Nuclear fusion and change in chromatin packing state in response to starvation in Triatoma infestans. Revta. Brasil. Genét. 1989, 12, 485–498. [Google Scholar]
- Andrade, C.G.T.J.; Mello, M.L.S. Phenotypes and number of Malpighian tubule nuclei in Triatoma infestans Klug along development and starvation. Revta. Brasil. Genét. 1987, 10, 449–457. [Google Scholar]
- Mello, M.L.S.; Pudney, M. Polyploidization of the BTC-32 cell line from Triatoma infestans (Hemiptera, Reduviidae). Genetica 1987, 74, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Wigglesworth, V.B. Polyploidy and nuclear fusion in the fat body of Rhodnius (Hemiptera). J. Cell Sci. 1967, 2, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Dolder, H.; Mello, M.L.S. Dados preliminares sobre partículas semelhantes a vírus em células de triatomíneos (Preliminary data on virus-like particles in triatomini cells). Revta. Saúde públ. 1978, 12, 104–109. [Google Scholar] [CrossRef]
- Dolder, H.; Mello, M.L.S. Virus-like particles in the Malpighian tubes of blood-sucking insects. Cell. Mol. Biol. 1978, 23, 299–310. [Google Scholar]
- Mello, M.L.S. Patterns of lability towards acid hydrolysis in heterochromatin and euchromatin of Triatoma infestans Klug. Cell. Mol. Biol. 1979, 24, 1–16. [Google Scholar]
- Alvarenga, E.M.; Mondin, M.; Martins, J.A.; Rodrigues, V.L.C.C.; Vidal, B.C.; Mello, M.L.S. Spatial distribution of AT- and GC-rich DNA within interphase cell nuclei of Triatoma infestans Klug. Micron 2011, 42, 568–578. [Google Scholar] [CrossRef]
- Schreiber, G.; Bogliolo, A.R.; Pinto, A.C. Cytogenetics of Triatominae: karyotype, DNA content, nuclear size and heteropyknosis of autosomes. Brazil. J. Biol. 1972, 32, 255–263. [Google Scholar]
- Solari, J. Autosomal synaptonemal complexes and sex chromosomes without axes in Triatoma infestans (Reduviidae, Hemiptera). Chromosoma 1979, 72, 225–240. [Google Scholar] [CrossRef]
- Morielle-Souza, A.; Azeredo-Oliveira, M.T.V. Differential characterization of holocentric chromosomes in triatomines (Heteroptera, Triatominae) using different staining techniques and fluorescent in situ hybridization. Genet. Mol. Res. 2007, 6, 713–720. [Google Scholar]
- Hirai, H.; Shomo, Y.; Arias, A.R.; Tada, I. Constitutive heterochromatin polymorphism of a Triatoma infestans strain, a main vector insect of Chagas disease. Jpn. J. Sant. Zool. 1991, 42, 301–303. [Google Scholar] [CrossRef]
- Panzera, F.; Perez, R.; Panzera, Y.; Ferrandis, I.; Ferreiro, M.J.; Calleros, L. Cytogenetics and genome evolution in the subfamily Triatominae (Hemiptera, Reduviidae). Cytogenet. Genome Res. 2010, 128, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Bardella, V.B.; da Rosa, J.A.; Vanzela, A.L.L. Origin and distribution of AT-rich repetitive DNA families in Triatoma infestans (Heteroptera). Infect. Genet. Evol. 2014, 23, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Imperador, C.H.L.; Bardella, V.B.; dos Anjos, E.H.; Rodrigues, V.L.C.C.; Cabral-de-Mello, D.C.; Mello, M.L.S. Spatial distribution of heterochromatin bodies in the nuclei of Triatoma infestans (Klug). Micr. Microanal. 2020, 26, 567–574. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. A conserved function for pericentromeric satellite DNA. ELife 2018, 7, e34122. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.G.T.J. Fenótipos nucleares com ênfase na heterocromatina, e frequências nucleares, em algumas espécies de triatomíneos. (Nuclear phenotypes with emphasis in heterochromatin, and nuclear frequencies in some triatomine species). Masters’ dissertation, University of Campinas, Campinas, 1984.
- Mello, M.L.S.; Vidal, B.C. Critical electrolyte concentration of the heterochromatin and euchromatin of Triatoma infestans. Cytobios 1989, 59, 87–93. [Google Scholar]
- Mayer, R.; Brero, A.; von Hase, J.; Schroeder, T.; Cremer, T.; Dietzel, S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005, 6, 44. [Google Scholar] [CrossRef]
- Jost, K.L.; Bertulat, B.; Rapp, A.; Brero, A.; Hardt, T.; Domaing, P.; Gösele, C.; Schulz, H.; Hübner, N.; Cardoso, M.C. Gene repositioning within the cell nucleus is not random and is determined by its genomic neighborhood. Epigenetics & Chromatin 2015, 8, 36. [Google Scholar]
- Poulet, A.; Duc, C.; Voisin, M.; Desset, S.; Tutois, S.; Vanrobays, E.; Benoit, M.; Evans, D.E.; Probst, A.V.; Tatout, C. The LINC complex contributes to heterochromatin organization and transcriptional gene silencing in plants. J. Cell Sci. 2017, 130, 590–601. [Google Scholar]
- Brändle, F.; Frühbauer, B.; Jagannathan, M. Principles and functions of pericentromeric satellite DNA clustering into chromocenters. Semin. Cell. Dev. Biol. 2022, 128, 26–39. [Google Scholar] [CrossRef]
- Imperador, C.H.L.; Rodrigues, V.L.C.C.; Mello, M.L.S. The topological distribution of the chromocenter in Panstrongylus megistus (Burmeister) Malpighian tubule cells examined by confocal microscopy. Cytologia 2021, 86, 47–54. [Google Scholar] [CrossRef]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nature Rev. 2018, 19, 229–244. [Google Scholar] [CrossRef]
- Bártova, E.; Krejci, J.; Harnicarová, A.; Galiová, G.; Kozubek, S. Histone modifications and nuclear architecture: A review. J. Histochem. Cytochem. 2008, 56, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.R.; Scalzo, D.; Groudine, M. Something silent this way forms: The functional organization of the repressive nuclear compartment. Annu. Rev. Cell Dev. Biol. 2013, 29, 241–270. [Google Scholar] [CrossRef] [PubMed]
- Mello, M.L.S.; Dolder, H.; Dias, C.A. Nuclear ultrastructure of Malpighian tubule cells in Triatoma infestans (Hemiptera, Reduviidae) under conditions of full nourishment and starvation. Brazil. J. Genet. 1990, 13, 5–17. [Google Scholar]
- Sandritter, W.; Kiefer, G.; Salm, R.; Moore, G.W.; Grimm, H.; Kiefer, R. DNA in heterochromatin cytophotometric pattern recognition image analysis among cell nuclei in duct epithelium and in carcinoma of the human breast. Beitr. Path. 1974, 151, 87–96. [Google Scholar] [CrossRef]
- Simões, L.C.G.; Cestari, A.N. Cromossomos politênicos: sistemas experimentais in vivo e in vitro. (Polytene chromosomes: in vivo and in vitro experimental systems). Ciênc, Cult. 1982, 34, 480–485. [Google Scholar]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. The modular mechanism of chromocenter formation in Drosophila. ELife 2019, 8, e43938. [Google Scholar] [CrossRef]
- Mello, M.L.S. Apoptosis in polyploid cells of the blood-sucking hemipteran, Triatoma infestans Klug. Caryologia 2005, 58, 281–287. [Google Scholar]
- Dantas, M.M.; Mello, M.L.S. Changes in the nuclear phenotypes of Triatoma infestans Klug induced by thermal shocks. Revta. Brasil. Genét. 1992, 15, 509–519. [Google Scholar]
- Rodrigues, V.L.C.C.; Mello, M.L.S.; Ferraz Filho, A.N.; Dantas, M.M. Sobrevivência e ocorrência de muda em Triatoma infestans Klug (Hemiptera, Reduviidae) após choque de temperatura (Survival and molting incidence in Triatoma infestans Klug (Hemiptera, Reduviidae) after temperature shocks). Revta. Saúde públ. 1991, 25, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Campos, S.G.P.; Rodrigues, V.L.C.C.; Wada, C.Y.; Mello, M.L.S. Effect of sequential cold shocks on survival and molting rate in Triatoma infestans Klug. Mem. Inst. Oswaldo Cruz 2002, 97, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.L.; Garcia, N.L.; Rodrigues, V.L.C.C.; Mello, M.L.S. Effect of sequential cold shocks on survival and molting incidence in Panstrongylus megistus (Burmeister) (Hemiptera, Reduviidae). Cryobiology 2001, 42, 74–77. [Google Scholar] [CrossRef]
- Campos, S.G.P.; Rodrigues, V.L.C.C.; Mello, M.L.S. Changes in nuclear phenotype frequencies following sequential cold shocks in Triatoma infestans (Hemiptera, Reduviidae). Mem. Inst. Oswaldo Cruz 2002, 97, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Mello, M.L.S.; Kubrusly, F.S.; Randi, M.A.F.; Rodrigues, V.L.C.C.; Ferraz Filho, A.N. Effect of heavy metals on chromatin supraorganization, nuclear phenotypes, and survival of Triatoma infestans. Entomol. Exp. Appl. 1995, 74, 209–218. [Google Scholar] [CrossRef]
- dos Anjos, E.H.M.; Rocha, M.A.; Vidal, B.C.; Mello, M.L.S. Exploration of DNA methylation in the chromatin of Triatoma infestans (Klug). Cytologia 2023, (in press). [Google Scholar]
- Bongiorni, S.; Cintio, O.; Prantera, C. The relationship between DNA methylation and chromosome imprinting in the coccid Planococcus citri. Genetics 1999, 151, 1471–1478. [Google Scholar] [CrossRef]
- Mandrioli, M.; Azzoni, P.; Lombardo, G.; Manicardi, G.C. Composition and epigenetic markers of heterochromatin in the aphid Aphis neri (Hemiptera: Aphididae). Cytogenet. Genome Res. 2011, 133, 67–77. [Google Scholar] [CrossRef]
- Alvarenga, E.M.; Rodrigues, V.L.C.C.; Moraes, A.S.; Naves, L.S.; Mondin, M.; Felisbino, M.B.; Mello, M.L.S. Histone epigenetic marks in heterochromatin and euchromatin of the Chagas disease vector, Triatoma infestans. Acta Histochem. 2016, 118, 401–412. [Google Scholar] [CrossRef]
- Wu, R.; Terry, A.V.; Singh, P.B.; Gilbert, D.M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell 2005, 16, 2872–2881. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profile of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Milavetz, B. Decoding the histone H4 lysine 20 methylation mark. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.A.; Vidal, B.C.; Mello, M.L.S. Sodium valproate modulates the methylation status of lysine residues 4, 9 and 27 in histone H3 of HeLa cells. Curr. Mol. Pharmacol. 2023, 16, 197–210. [Google Scholar] [PubMed]
- Rice, J.C.; Briggs, S.D.; Ueberheide, B.; Barber, C.M.; Shabanowitz, J.; Hunt, D.F.; Shinkai, Y.; Allis, C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 2003, 12, 1591–1598. [Google Scholar] [CrossRef]
- Suka, N.; Luo, K.; Grunstein, M. Sir2p and Sas2p opposingly regulate acetylation of histone H4 lysine 16 and spreading of heterochromatin. Nature Genet. 2002, 32, 378–383. [Google Scholar] [CrossRef]
- Robinson, P.J.J.; Na, W.; Routh, A.; Martino, F.; Chapman, L.; Roeder, R.G.; Rhodes, D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 2008, 381, 816–825. [Google Scholar] [CrossRef]
- Zhou, Y.; Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005, 15, 1434–1438. [Google Scholar] [CrossRef]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Milavetz, B. Decoding the histone H4 lysine 20 methylation mark. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 440–452. [Google Scholar] [CrossRef]
- Corvalan, A.Z.; Coller, H.A. Methylation of histone 4’s lysine 20: a critical analysis of the state of the field. Physiol. Genomics 2021, 53, 22–32. [Google Scholar] [CrossRef]
- Göttlicher, M.; Minucci, S.; Zhu, P.; Kramer, O.H.; Schinpf, A.; Giavara, S.; Sleeman, J.P.; LoCoco, F.; Nervi, C.; Pelicci, P.G.; Heinzel, T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001, 20, 6969–6978. [Google Scholar] [CrossRef]
- Phiel, C.J.; Zhang, F.; Huang, E.Y.; Guenther, M.G.; Lazar, M.A.; Klein, P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvu lsant, mood stabilizer, and teratogene. J. Biol. Chem. 2001, 276, 36734–36741. [Google Scholar] [CrossRef]
- Detich, N.; Bovenzi, V.; Szyf, M. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 2003, 278, 27586–27592. [Google Scholar] [CrossRef]
- Marchion, D.C.; Bicaku, E.; Daud, A.I.; Sullivan, D.M.; Munster, P.N. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005, 65, 3815–3822. [Google Scholar] [CrossRef] [PubMed]
- Felisbino, M.B.; Tamashiro, W.M.S.C.; Mello, M.L.S. Chromatin remodeling, cell proliferation and cell death in valproico acid-treated HeLa cells. PLoS ONE 2011, 6, e29144. [Google Scholar] [CrossRef] [PubMed]
- Felisbino, M.B.; Gatti, M.S.V.; Mello, M.L.S. Changes in chromatin structure in NIH 3T3 cells induced by valproic acid and trichostatin A. J. Cell Physiol. 2014, 115, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.A.; Veronezi, G.M.B.; Felisbino, M.B.; Gatti, M.S.V.; Tamashiro, W.M.S.C.; Mello, M.L.S. Sodium valproate and 5-aza-2’-deoxycytidine differentially modulate DNA demethylation in G1 phase-arrested and proliferative HeLa cells. Sci. Rep. 2019, 9, 18236. [Google Scholar] [CrossRef]
- Sargolzaei, J.; Rabbani-Chadegani, A.; Mollaei, H.; Deezagi, A. Spectroscopic analysis of the interaction of valproic acid with histone H1 in solution and in chromatin structure. Int. J. Biol. Macromol. 2017, 99, 427–432. [Google Scholar] [CrossRef]
- Vidal, B.C.; Mello, M.L.S. Sodium valproate (VPA) interactions eith DNA and histones. Int. J. Biol. Macromol. 2020, 163, 219–231. [Google Scholar] [CrossRef]
- Baumann, C.; Zhang, Z.; Zhu, L.; Fan, Y.; De La Fuente, R. Changes in chromatin accessibility lasndscape and histone H3 core acetylation during valproic acid-induced differentiation of embryonic stem cells. Epigenetics & Chromatin 2021, 14, 58. [Google Scholar]
- Mello, M.L.S. Sodium valproate-induced chromatin remodeling. Front. Cell Dev. Biol. 2021, 9, 645518. [Google Scholar] [CrossRef]
- Duenas-Gonzalez, A.; Candelaria, M.; Perez-Plascencia, C.; Perez-Cardenas, E.; de la Cruz-Hernandez, E.; Herrera, L.A. Valproic acid as epigenetic cancer drug: Preclinical, clinical and transcriptional effects on solid tumors. Cancer Treat. Rev. 2008, 34, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Chateauvieux, S.; Morceau, F.; Dicato, M.; Diederich, M. Molecular and therapeutical potential and toxicity of valproic acid. J. Biomed. Biotechnol. 2010, 2010, 479364. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Coy, D.H. Anti-convulsant drug valproic acid in cancers and in combination anti-cancer therapeutics. Mod. Chem. Appl. 2014, 2, 1000118. [Google Scholar]
- Heers, H.; Stanislaw, J.; Harrelson, J.; Lee, M.W. Valproic acid as an adjunctive therapeutic agent for the treatment of breast cancer. Eur. J. Pharmacol. 2018, 835, 61–74. [Google Scholar] [CrossRef]
- Santos, D.S.; Rocha, M.A.; Mello, M.L.S. Epigenetic studies in insects and the valproic acid perspective. Brazil. J. Biol. 2022, 84, e256045. [Google Scholar] [CrossRef]
- Bassani, A.; Rocha, M.A.; Rodrigues, V.L.C.C.; Santos, D.S.; Nascimento, J.D.; da Rosa, J.A.; Mello, M.L.S. Effects of sodium valproate on the chromatin of Triatoma infestans (Klug, 1834) (Hemiptera, Reduviidae) under in vitro culture conditions. Acta Histochem. 2021, 123, 151695. [Google Scholar] [CrossRef]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Amassalha, L.M.; Bakman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 2018, 29, 220–233. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).