Submitted:
30 June 2023
Posted:
04 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Methods
2.3. Synthesis
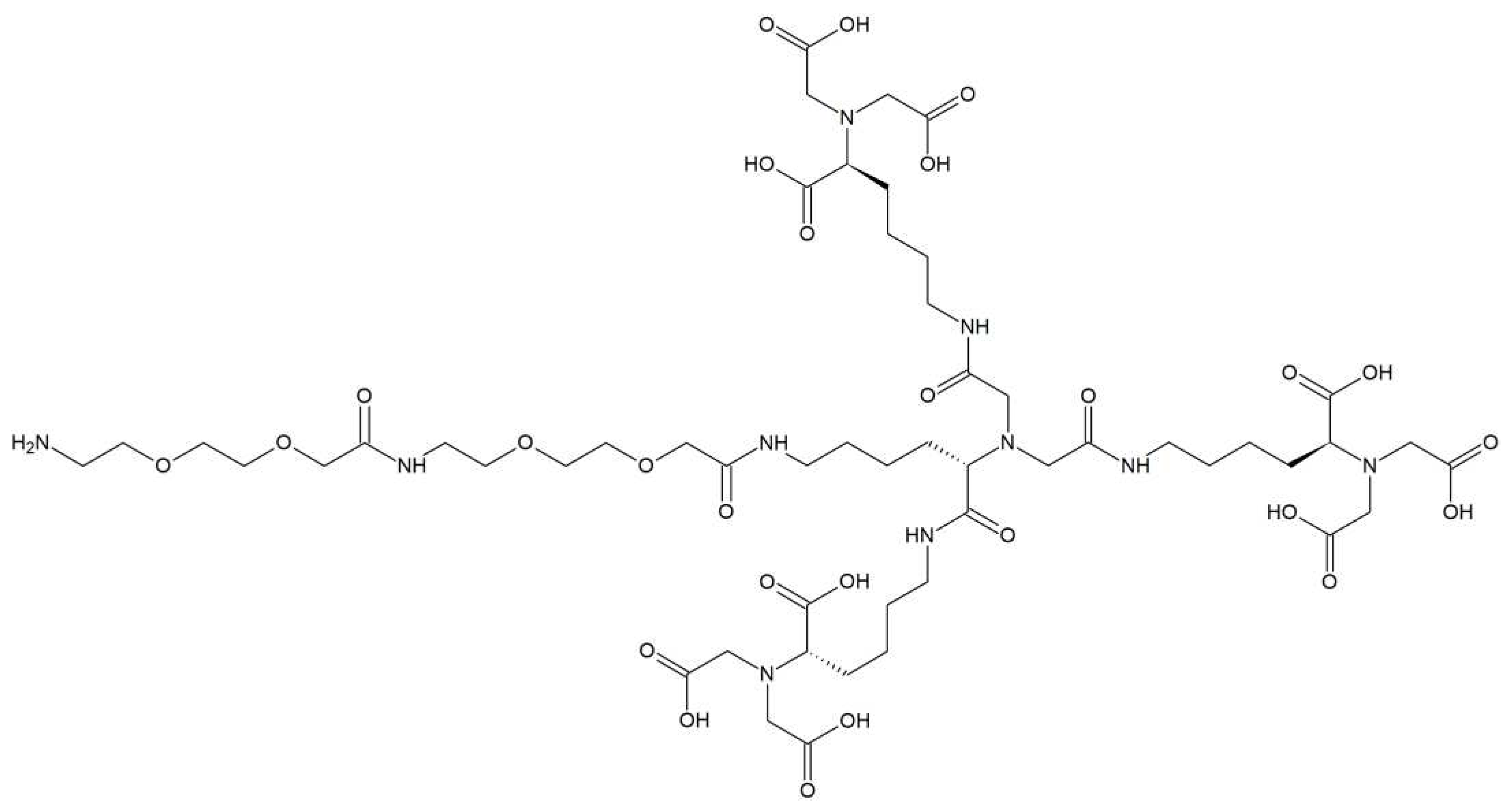
2.3.1. Synthesis of NTA derivatives
2.3.2. Synthesis of monomers, polymers and polymer/protein complexes
2.4. Cells and Media
2.5. Flow Cytometry
2.6. Confocal microscopy
3. Results and Discussion
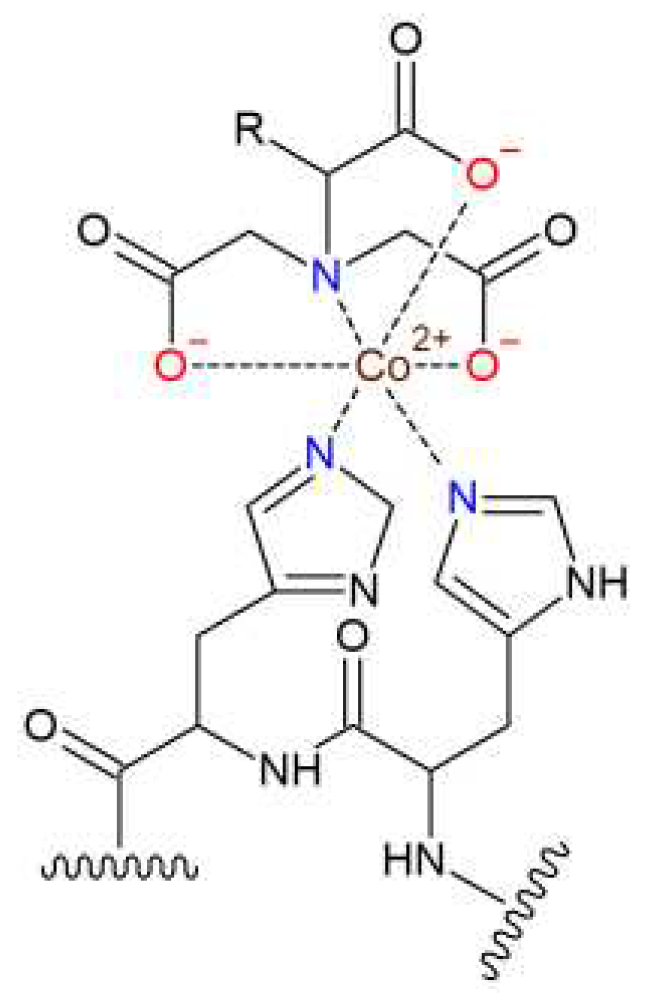
3.1. Synthesis of tris-NTA chelator
3.2. Synthesis of polymers and polymer/protein complexes
3.3. Preparation and characterization of the polymer/protein complexes
3.3.1. SEC analysis
3.3.2. DLS analysis
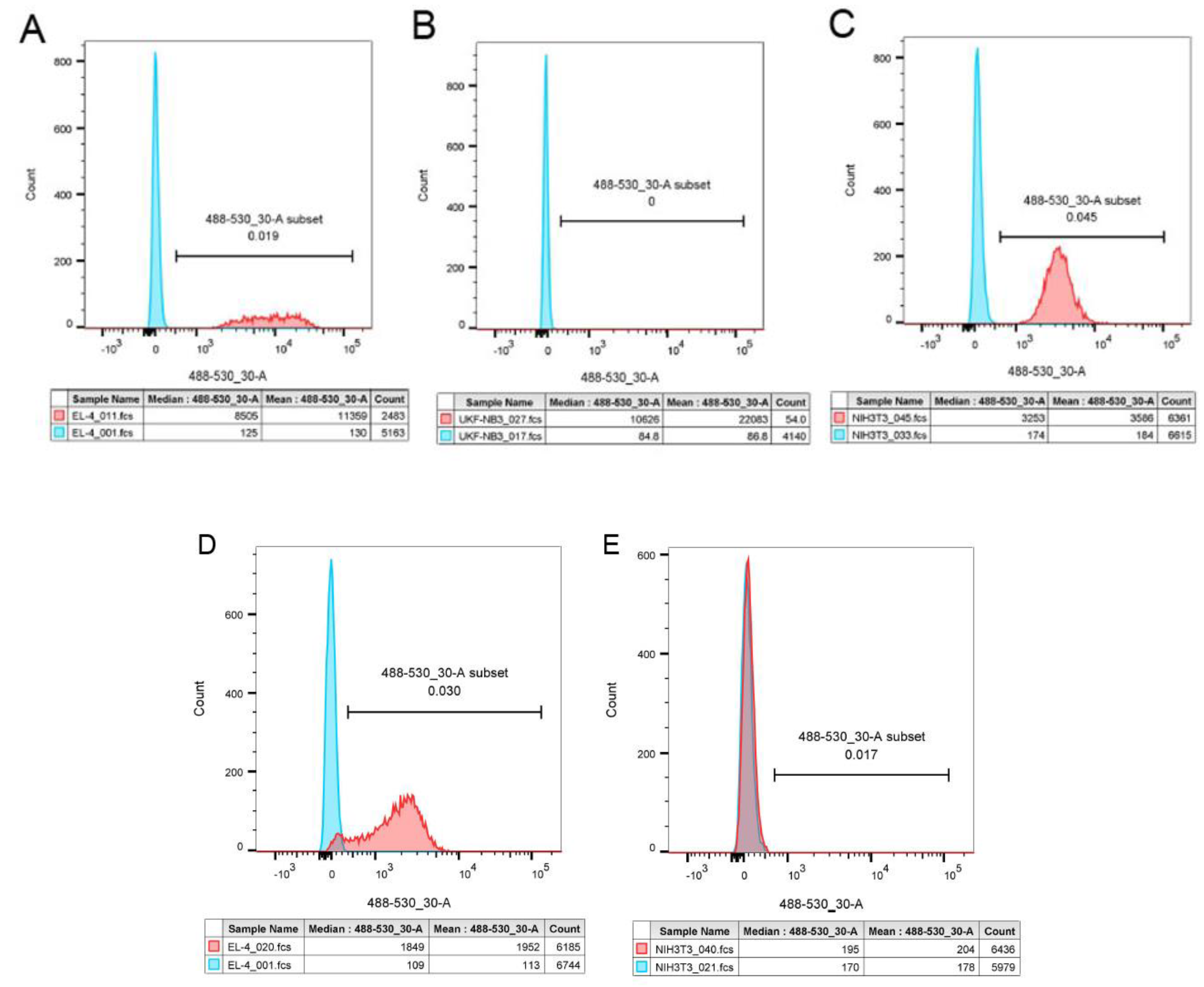
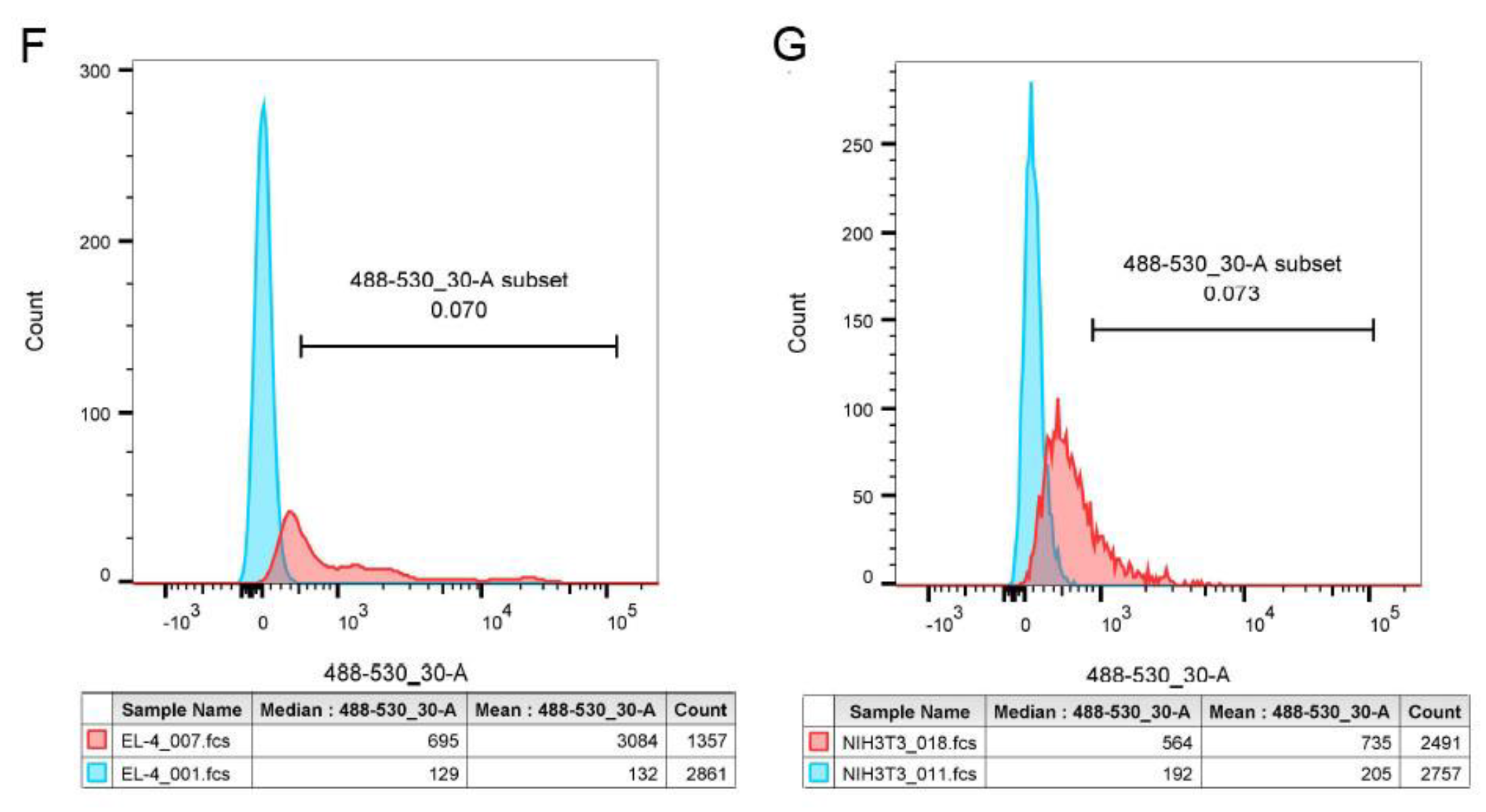
3.3.2. Flow cytometry analysis
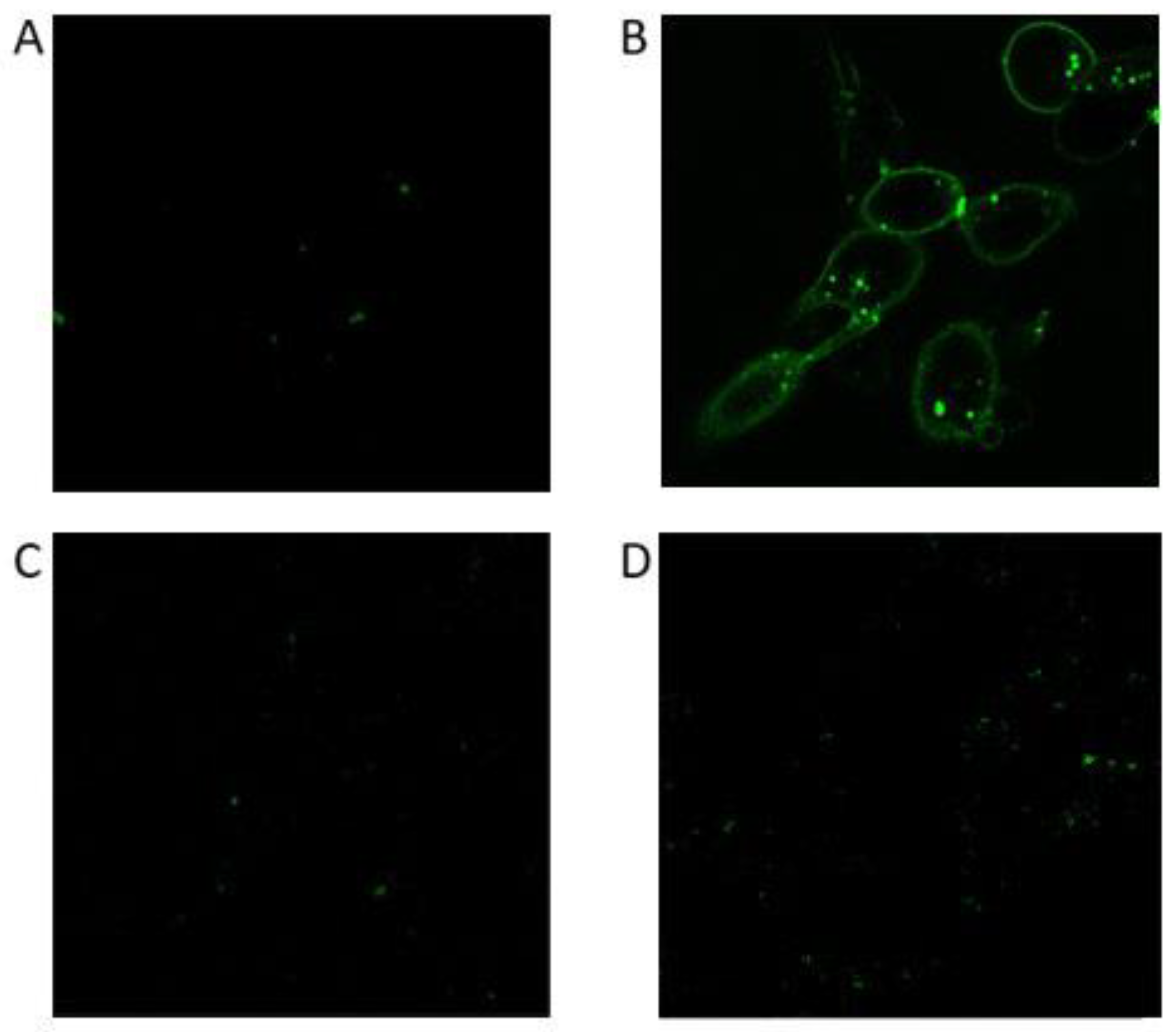
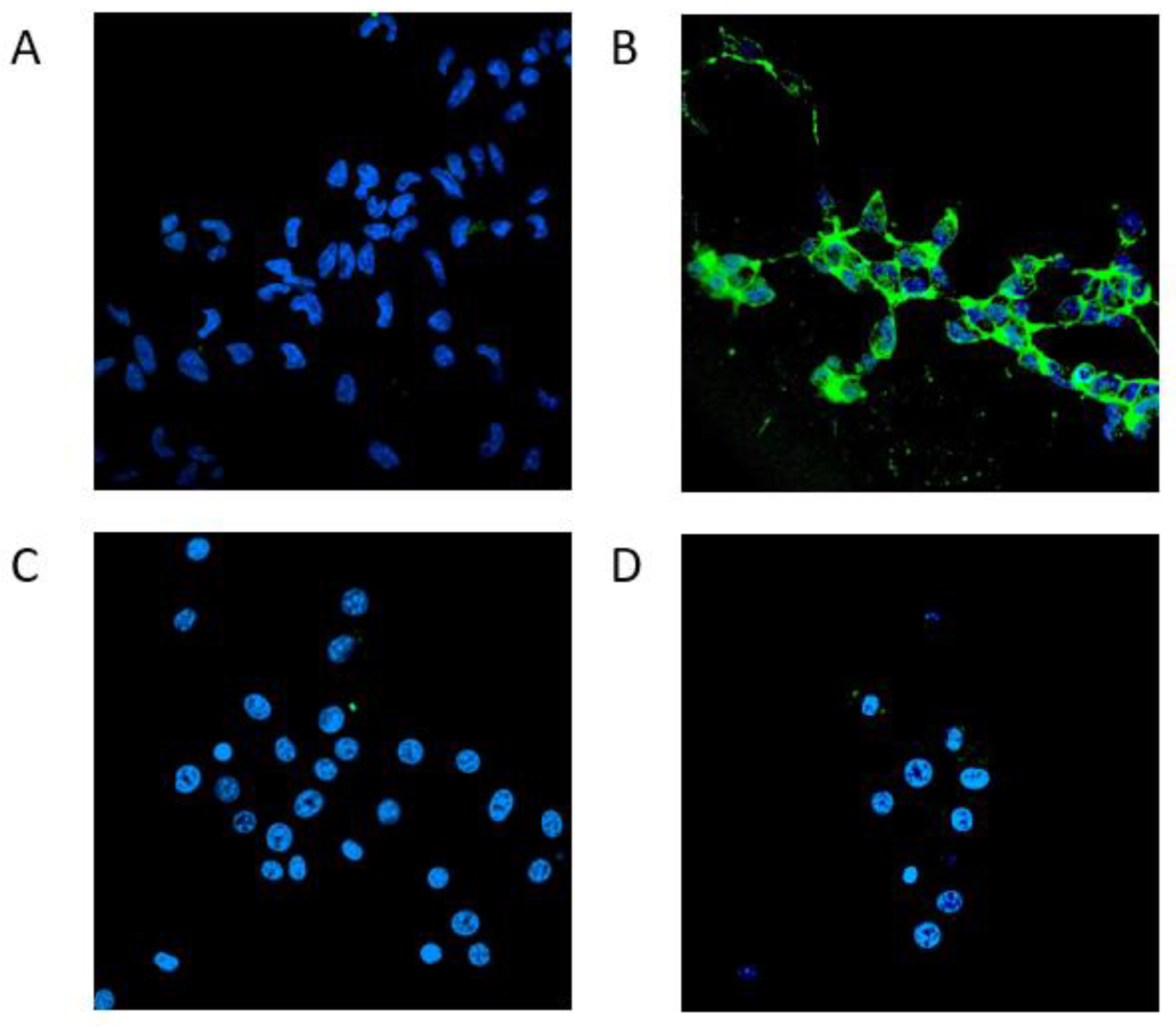
3.3.2. Confocal microscopy analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kopeček, J.; Yang, J. Polymer nanomedicines. Adv. Drug Deliv. Rev. 2020, 156, 40–64. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.-W.; Kopeček, J. Drug-free macromolecular therapeutics – a new paradigm in polymeric nanomedicines. Biomater. Sci. 2015, 3, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Antos, J.M.; Truttmann, M.C.; Ploegh, H.L. Recent advances in sortase-catalyzed ligation methodology. Curr. Opin. Struct. Biol. 2016, 38, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Böker, A.; Glebe, U. Broadening the scope of sortagging. RSC Adv. 2019, 9, 4700–4721. [Google Scholar] [CrossRef] [PubMed]
- Laga, R.; Pola, R.; Ulbrich, K.; Hořejší, M.; Sieglová, I.; Král, V.; Fábry, M.; Pechar, M. Avidin-conjugated polymers with monobiotinylated antibody fragments: A new strategy for the noncovalent attachment of recombinant proteins for polymer therapeutics. J. Bioact. Compat. Polym. 2013, 28, 289–299. [Google Scholar] [CrossRef]
- Wu, K.; Yang, J.; Liu, J.; Kopeček, J. Coiled-coil based drug-free macromolecular therapeutics: In vivo efficacy. J. Control. Release 2012, 157, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Pechar, M.; Pola, R.; Janouskova, O.; Sieglova, I.; Kral, V.; Fabry, M.; Tomalova, B.; Kovar, M.; Janoušková, O.; Sieglová, I.; et al. Polymer Cancerostatics Targeted with an Antibody Fragment Bound via a Coiled Coil Motif: In Vivo Therapeutic Efficacy against Murine BCL1 Leukemia. Macromol. Biosci. 2018, 18, 1700173. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.-W.; Yang, J.; Zhang, R.; Sima, M.; Kopeček, J. Cell Surface Self-Assembly of Hybrid Nanoconjugates via Oligonucleotide Hybridization Induces Apoptosis. ACS Nano 2014, 8, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Spriestersbach, A.; Kubicek, J.; Schäfer, F.; Block, H.; Maertens, B. Purification of His-Tagged Proteins. In Methods in Enzymology; Academic Press, 2015; Vol. 559, pp. 1–15.
- Beranová, J.; Knedlík, T.; Šimková, A.; Šubr, V.; Kostka, L.; Etrych, T.; Šácha, P.; Konvalinka, J. Tris-(nitrilotriacetic acid)-decorated polymer conjugates as tools for immobilization and visualization of his-tagged proteins. Catalysts 2019. [Google Scholar] [CrossRef]
- Pola, R.; Král, V.; Filippov, S.K.; Kaberov, L.; Etrych, T.; Sieglová, I.; Sedláček, J.; Fábry, M.; Pechar, M. Polymer Cancerostatics Targeted by Recombinant Antibody Fragments to GD2-Positive Tumor Cells. Biomacromolecules 2019, 20, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K. V; Saarinen, U.M.; Neely, J.E.; Landmeier, B.; Donovan, D.; Coccia, P.F. Monoclonal antibodies to a glycolipid antigen on human neuroblastoma cells. Cancer Res 1985, 45, 2642–2649. [Google Scholar] [PubMed]
- Chang, H.R.; Cordon-Cardo, C.; Houghton, A.N.; Cheung, N.K. V; Brennan, M.F. Expression of disialogangliosides GD2 and GD3 on human soft tissue sarcomas. Cancer 1992, 70, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.C.; Kostakoglu, L.; Kris, M.G.; Yeh, S.D.J.; Larson, S.M.; Finn, R.D.; Oettgen, H.F.; Cheung, N.-K. V. Targeting of small-cell lung cancer using the anti-GD2 ganglioside monoclonal antibody 3F8: A pilot trial. Eur. J. Nucl. Med. 1996, 23, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, K.; Šubr, V.; Strohalm, J.; Plocová, D.; Jelı́nková, M.; Řı́hová, B. Polymeric drugs based on conjugates of synthetic and natural macromolecules. J. Control. Release 2000, 64, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Šubr, V.; Ulbrich, K. Synthesis and properties of new N-(2-hydroxypropyl)methacrylamide copolymers containing thiazolidine-2-thione reactive groups. React. Funct. Polym. 2006, 66, 1525–1538. [Google Scholar] [CrossRef]
- Huang, Z.; Hwang, P.; Watson, D.S.; Cao, L.; Szoka, F.C. Tris-nitrilotriacetic acids of subnanomolar affinity toward hexahistidine tagged molecules. Bioconjug. Chem. 2009, 20, 1667–1672. [Google Scholar] [CrossRef] [PubMed]
- Kokufuta, M.K.; Sato, S.; Kokufuta, E. LCST behavior of copolymers of N-isopropylacrylamide and N-isopropylmethacrylamide in water. Colloid Polym. Sci. 2012, 290, 1671–1681. [Google Scholar] [CrossRef]
- Kuperkar, K.; Patel, D.; Atanase, L.I.; Bahadur, P. Amphiphilic Block Copolymers: Their Structures, and Self-Assembly to Polymeric Micelles and Polymersomes as Drug Delivery Vehicles. Polymers (Basel). 2022, 14, 4702. [Google Scholar] [CrossRef] [PubMed]
- Tavares, M.R.; Pechar, M.; Chytil, P.; Etrych, T. Polymer-Based Drug-Free Therapeutics for Anticancer, Anti-Inflammatory, and Antibacterial Treatment. Macromol. Biosci. 2021, 21. [Google Scholar] [CrossRef] [PubMed]
- Gambles, M.T.; Li, J.; Wang, J.; Sborov, D.; Yang, J.; Kopeček, J. Crosslinking of CD38 Receptors Triggers Apoptosis of Malignant B Cells. Mol. 2021, Vol. 26, Page 4658 2021, 26, 4658. [Google Scholar] [CrossRef] [PubMed]
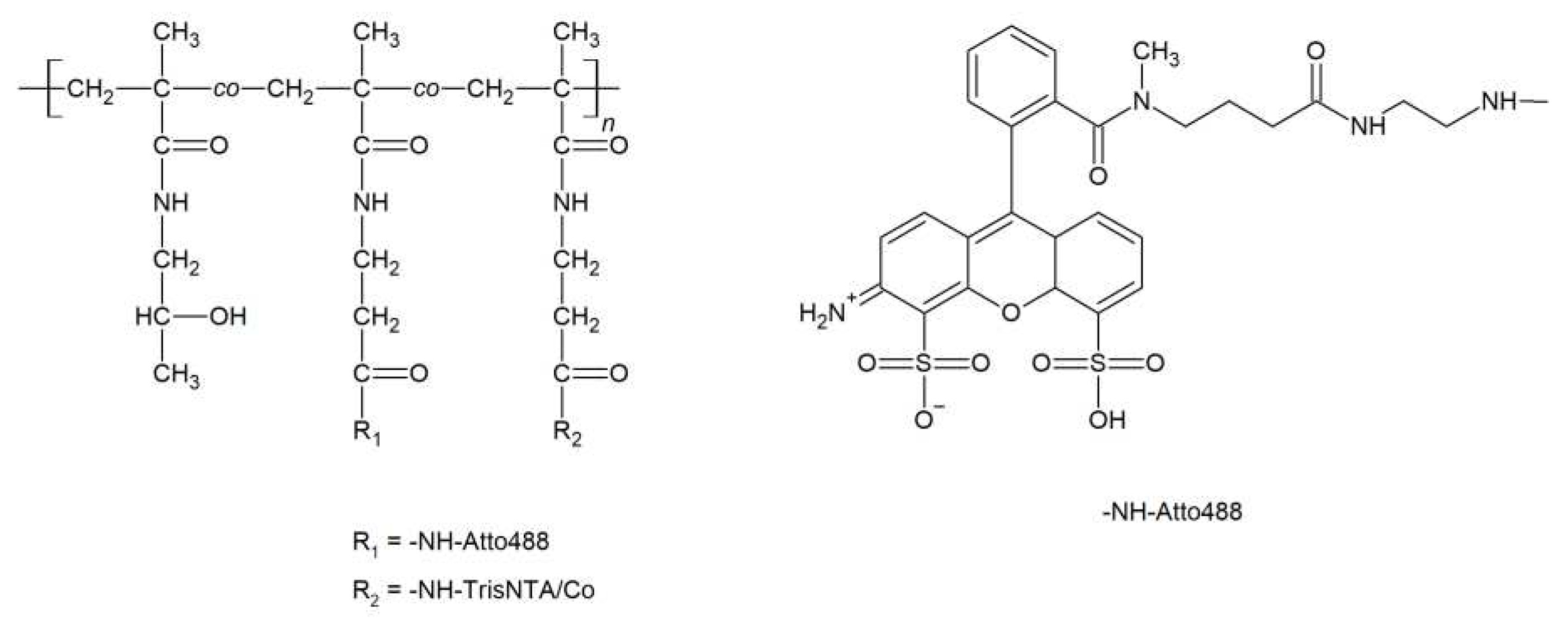
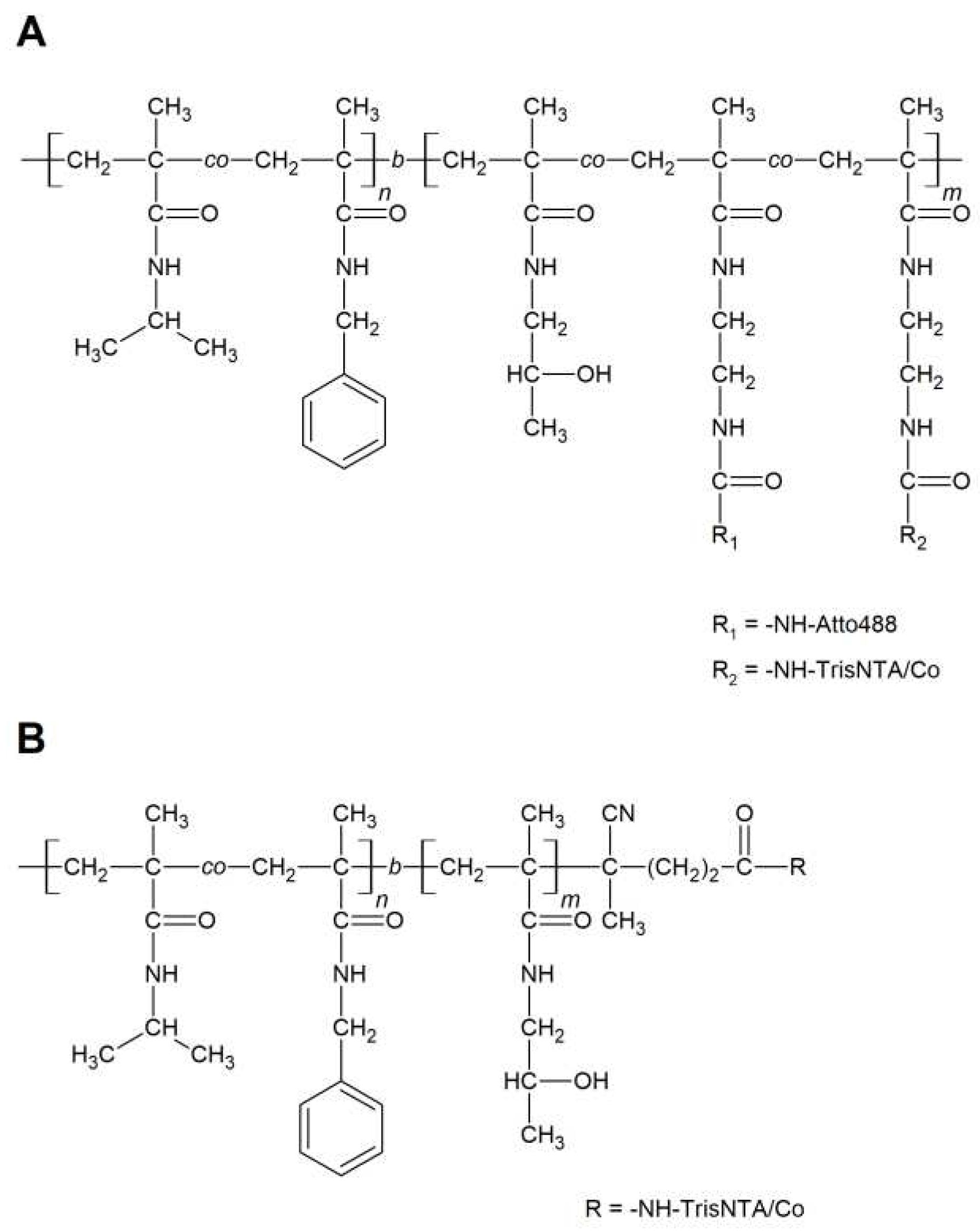
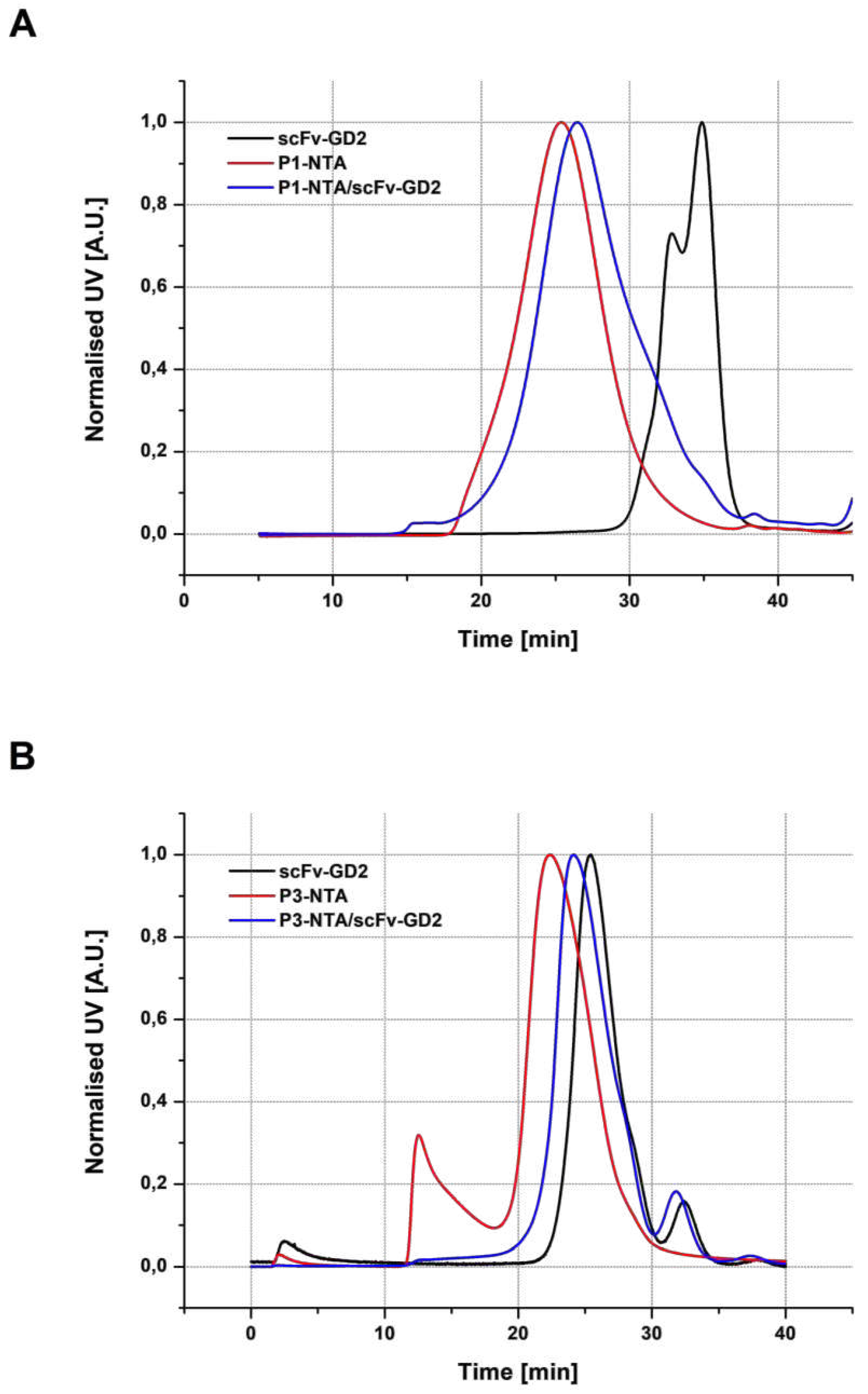
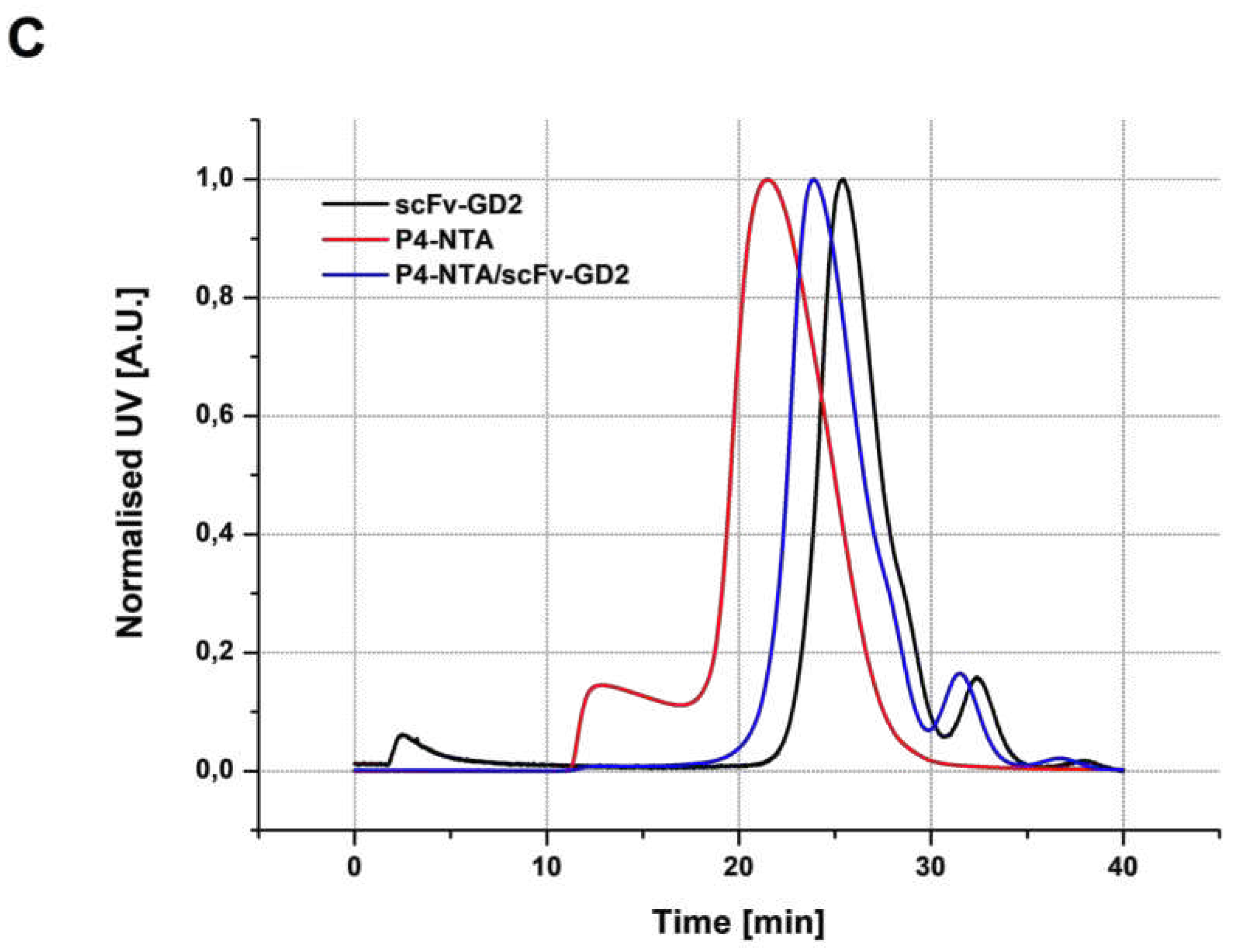

| Sample | Structure | Mw [kg·mol−1] | Ð |
|---|---|---|---|
| P1-TT | P(HPMA-co-Ma-AP-TT) | 62.0 | 1.05 |
| P1-NTA | P(HPMA-co-Ma-AP-Atto488--co-Ma-AP-trisNTA-Co) | 113.0 | 1.20 |
| P1-NTA/scFv-GD2 | P(HPMA-co-Ma-AP-Atto488--co-Ma-AP-trisNTA-Co/scFv-GD2) | 168.0 | 1.37 |
| P2 | P(NIPMAM-co-BnMAM)-DTB | 10.9 | 1.08 |
| P3 | P(NIPMAM-co-BnMAM)-b--P(HPMA-co-Ma-AP-TT)-DTB | 20.5 | 1.09 |
| P3-TT | P(NIPMAM-co-BnMAM)-b--P(HPMA-co-Ma-AP-TT)-IBN | 21.6 | 1.09 |
| P3-NTA | P(NIPMAM-co-BnMAM)-b--P(HPMA-co-Ma-AP-trisNTA-Co--co-Ma-AP-Atto488)]-IBN | 23.8 | 1.08 |
| P3-NTA/scFv-GD2 | P(NIPMAM-co-BnMAM)-b--P(HPMA-co-Ma-AP-trisNTA-Co/scFv-GD2-co-Ma-AP-Atto488)]-IBN | n.d. | n.d. |
| P4 | P(NIPMAM-co-BnMAM)-b--P(HPMA)-DTB | 36.1 | 1.04 |
| P4-TT | P(NIPMAM-co-BnMAM)-b--P(HPMA)-TT | 36.8 | 1.05 |
| P4-NTA | P(NIPMAM-co-BnMAM)-b-P(HPMA)--TrisNTA-Co | 41.4 | 1.08 |
| P4-NTA/scFv-GD2 | P(NIPMAM-co-BnMAM)-b-P(HPMA)--TrisNTA-Co/scFv-GD2 | n.d. | n.d. |
| Sample | RH15 °C [nm] | RH37 °C [nm] | PDI37 °C |
|---|---|---|---|
| scFv-GD2 | 2.9 ± 0.3 | 2.9 ± 0.2 | n.d. |
| P3-NTA | 2.9 ± 0.5 | 23.8 ± 0.7 | 0.175 ± 0.015 |
| P3-NTA/scFv-GD2 | 2.8 ± 0.4 | 27.1 ± 1.4 | 0.254 ± 0.009 |
| P4-NTA | 5.3 ± 0.8 | 17.4 ± 0.6 | 0.206 ± 0.019 |
| P4-NTA/scFv-GD2 | 3.9 ± 0.6 | 19.1 ± 0.7 | 0.213 ± 0.029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





