Submitted:
28 June 2023
Posted:
29 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and methods
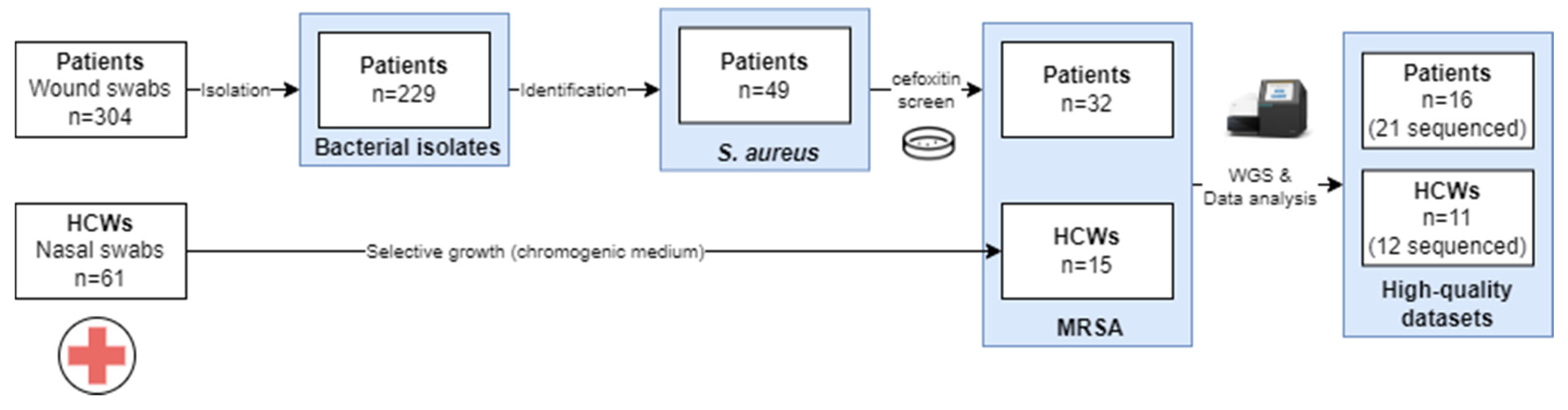
2.1. Study design
2.2. Sample collection
2.3. Bacterial identification
2.4. Whole genome sequencing
2.5. Quality control and preprocessing
2.6. Isolate characterization
2.7. Phylogenomic investigation
2.8. Ethics approval and consent to participate.
3. Results
3.1. Sample collection and species identification
3.2. WGS data analysis
Preprocessing and data quality evaluation
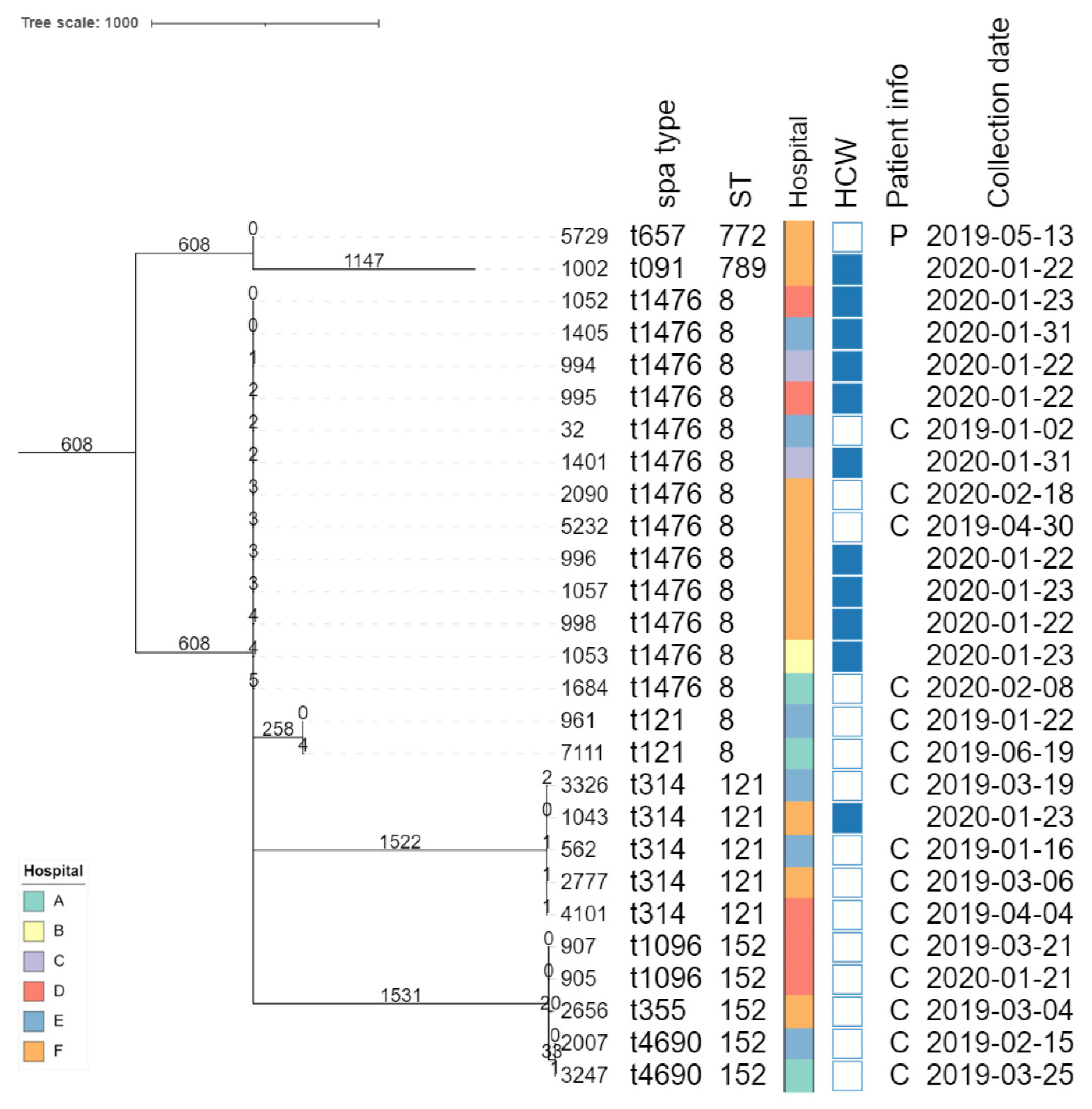
Isolate typing
AMR prediction
Virulence gene detection
Phylogenomic investigation
4. Discussion
5. Conclusion
Funding
Supplementary materials
Author’s contributions
Data availability statement
Acknowledgements
Competing interests
References
- Kondo S, Phokhaphan P, Tongsima S, Ngamphiw C, Phornsiricharoenphant W, Ruangchai W, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus genotype ST764-SCCmec type II in Thailand. Sci Rep. Nature Research; 2022;12.
- Ranklin F, Owy DL. Review Articles Medical Progress. 1998.
- Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y, et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother. 2013; 45:117–36.
- Achek R, El-Adawy H, Hotzel H, Hendam A, Tomaso H, Ehricht R, et al. Molecular characterization of staphylococcus aureus isolated from human and food samples in northern algeria. Pathogens. 2021; 10:1–19.
- Allegranzi B, Nejad SB, Combescure C, Graafmans W, Attar H, Donaldson L, et al. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. The Lancet. 2011; 377:228–41.
- Kim DH, Spencer M, Davidson SM, Li L, Shaw JD, Gulczynski D, et al. Institutional prescreening for detection and eradication of methicillin-resistant Staphylococcus aureus in patients undergoing elective orthopaedic surgery. Journal of Bone and Joint Surgery. Journal of Bone and Joint Surgery Inc.; 2010; 92:1820–6.
- Roy S, Patkar A, Daskiran M, Levine R, Hinoul P, Nigam S. Clinical and economic burden of surgical site infection in hysterectomy. Surg Infect (Larchmt). Mary Ann Liebert Inc.; 2014; 15:266–73.
- Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. Journal of Hospital Infection [Internet]. The Hospital Infection Society; 2008; 70:3–10. Available from. [CrossRef]
- Upreti N, Rayamajhee B, Sherchan SP, Choudhari MK, Banjara MR. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum β-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal 11 Medical and Health Sciences 1103 Clinical Sci. Antimicrob Resist Infect Control. Antimicrobial Resistance & Infection Control; 2018;7:1–10.
- Benito D, Gómez P, Aspiroz C, Zarazaga M, Lozano C, Torres C. Molecular characterization of Staphylococcus aureus isolated from humans related to a livestock farm in Spain, with detection of MRSA-CC130 carrying mecC gene: A zoonotic case? Enferm Infecc Microbiol Clin. 2016;34:280–5.
- Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, et al. SCC mec Finder, a Web-Based Tool for Typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus Using Whole-Genome Sequence Data. mSphere. American Society for Microbiology; 2018;3.
- Andrade MM, Luiz WB, da Silva Oliveira Souza R, Amorim JH. The History of Methicillin-Resistant Staphylococcus aureus in Brazil. Canadian Journal of Infectious Diseases and Medical Microbiology. Hindawi Limited; 2020.
- Ouedraogo AS, Dunyach-Remy C, Kissou A, Sanou S, Poda A, Kyelem CG, et al. High nasal carriage rate of Staphylococcus aureus containing panton-valentine leukocidin- and EDIN-encoding genes in community and hospital settings in Burkina Faso. Front Microbiol. Frontiers Media S.A.; 2016;7.
- Kondo S, Phokhaphan P, Tongsima S, Ngamphiw C, Phornsiricharoenphant W, Ruangchai W, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus genotype ST764-SCCmec type II in Thailand. Sci Rep [Internet]. Nature Publishing Group UK; 2022;12:1–8. Available from. [CrossRef]
- Bosch T, Pluister GN, van Luit M, Landman F, van Santen-Verheuvel M, Schot C, et al. Multiple-locus variable number tandem repeat analysis is superior to spa typing and sufficient to characterize MRSA for surveillance purposes. Future Microbiol. 2015;10:1155–62.
- El-Adawy H, Ahmed M, Hotzel H, Monecke S, Schulz J, Hartung J, et al. Characterization of methicillin-resistant staphylococcus aureus isolated from healthy Turkeys and broilers using DNA microarrays. Front Microbiol. 2016;7.
- Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, et al. Meticillin-resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents [Internet]. Elsevier B.V.; 2012;39:273–82. Available from. [CrossRef]
- Wong JWH, Ip M, Tang A, Wei VWI, Wong SYS, Riley S, et al. Prevalence and risk factors of community-associated methicillin-resistant staphylococcus aureus carriage in asia-pacific region from 2000 to 2016: A systematic review and meta-analysis. Clin Epidemiol. Dove Medical Press Ltd; 2018. p. 1489–501.
- Enany S, Yaoita E, Yoshida Y, Enany M, Yamamoto T. Molecular characterization of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol Res [Internet]. Elsevier; 2010;165:152–62. Available from. [CrossRef]
- Mariem BJJ, Ito T, Zhang M, Jin J, Li S, Ilhem BBB, et al. Molecular characterization of methicillin-resistant Panton-valentine leukocidin positive staphylococcus aureus clones disseminating in Tunisian hospitals and in the community. BMC Microbiol. 2013;13.
- Antri K, Rouzic N, Dauwalder O, Boubekri I, Bes M, Lina G, et al. High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clinical Microbiology and Infection [Internet]. European Society of Clinical Infectious Diseases; 2011;17:526–32. Available from. [CrossRef]
- Eiff, V. Nasal Carriage As a Sourc E of Sta Ph Yloc Occ Us Aureus Bac Ter Emia Nasal Carriage As a Source of Staphylococcus Aureus Bacteremia. English Journal. 2001;344:11–6.
- Yehouenou CL, Dohou AM, Fiogbe AD, Esse M, Degbey C, Simon A, et al. Hand hygiene in surgery in Benin : opportunities and challenges. 2020;1–8.
- Borchardt RA, Tzizik D. Update on surgical site infections: The new CDC guidelines. J Am Acad Physician Assist. Lippincott Williams and Wilkins; 2018;31:52–4.
- Yehouenou CL, Kpangon AA, Affolabi D, Rodriguez-Villalobos H, van Bambeke F, Dalleur O, et al. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Benin. Ann Clin Microbiol Antimicrob. BioMed Central Ltd; 2020;19.
- Karmakar A, Dua P, Ghosh C. Biochemical and Molecular Analysis of Staphylococcus aureus Clinical Isolates from Hospitalized Patients. Canadian Journal of Infectious Diseases and Medical Microbiology. Hindawi Limited; 2016;2016.
- Cefoxitin disk MRSA.
- Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clinical Microbiology and Infection. Blackwell Publishing Ltd; 2010;16:1614–9.
- Unal S, Hoskins J, Flokowitsch,’ JE, Wu CYE, Preston DA, Skatrudl PL. Detection of Methicillin-Resistant Staphylococci by Using the Polymerase Chain Reaction. J Clin Microbiol. 1992.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. Oxford University Press; 2014;30:2114–20.
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo Assembler. Curr Protoc Bioinformatics. John Wiley and Sons Inc.; 2020;70.
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. BioMed Central Ltd.; 2019;20.
- Low AJ, Koziol AG, Manninger PA, Blais B, Carrillo CD. ConFindr: Rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ. PeerJ Inc.; 2019;2019.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10.
- Bogaerts B, Nouws S, Verhaegen B, Denayer S, van Braekel J, Winand R, et al. Validation strategy of a bioinformatics whole genome sequencing workflow for Shiga toxin-producing Escherichia coli using a reference collection extensively characterized with conventional methods. Microb Genom. Microbiology Society; 2021;7.
- Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. Nature Research; 2021;11.
- Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. American Society for Microbiology; 2014;52:1501–10.
- Ren Y, Chakraborty T, Doijad S, Falgenhauer L, Falgenhauer J, Goesmann A, et al. Prediction of antimicrobial resistance based on whole-genome sequencing and machine learning. Bioinformatics. Oxford University Press; 2022;38:325–34.
- Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. Grapetree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. Cold Spring Harbor Laboratory Press; 2018;28:1395–404.
- Dallman T, Ashton P, Schafer U, Jironkin A, Painset A, Shaaban S, et al. SnapperDB: a database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. Oxford University Press; 2018;34:3028–9.
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. Oxford University Press; 2016;44:W16–21.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. Oxford University Press; 2018;35:1547–9.
- Ouedraogo AS, Dunyach-Remy C, Kissou A, Sanou S, Poda A, Kyelem CG, et al. High nasal carriage rate of Staphylococcus aureus containing panton-valentine leukocidin- and EDIN-encoding genes in community and hospital settings in Burkina Faso. Front Microbiol. Frontiers Media S.A.; 2016;7.
- Kesah C, Redjeb S ben, Odugbemi TO, Boye CSB, Dosso M, Ndinya Achola JO, et al. Prevalence of methicillin-resistant Staphylococcus aureus in eight African hospitals and Malta. Clinical Microbiology and Infection [Internet]. European Society of Clinical Infectious Diseases; 2003;9:153–6. Available from. [CrossRef]
- Antri K, Rouzic N, Dauwalder O, Boubekri I, Bes M, Lina G, et al. High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clinical Microbiology and Infection [Internet]. European Society of Clinical Infectious Diseases; 2011;17:526–32. Available from. [CrossRef]
- Alioua MA, Labid A, Amoura K, Bertine M, Gacemi-Kirane D, Dekhil M. Emergence of the European ST80 clone of community-associated methicillin-resistant Staphylococcus aureus as a cause of healthcare-associated infections in Eastern Algeria. Med Mal Infect [Internet]. Elsevier Masson SAS; 2014;44:180–3. Available from. [CrossRef]
- Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, et al. Prediction of staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol. American Society for Microbiology; 2014;52:1182–91.
- Wolters M, Frickmann H, Christner M, Both A, Rohde H, Oppong K, et al. Molecular characterization of staphylococcus aureus isolated from chronic infected wounds in rural Ghana. Microorganisms. 2020;8:1–7.
- Akinkunmi EO, Lamikanra A. Species distribution and antibiotic resistance in coagulase-negative staphylococci colonizing the gastrointestinal tract of children in Ile-Ife, Nigeria. Tropical Journal of Pharmaceutical Research. 2010;9:35–43.
- Schaumburg F, Alabi AS, Peters G, Becker K. New epidemiology of Staphylococcus aureus infection in Africa. Clinical Microbiology and Infection [Internet]. European Society of Clinical Infectious Diseases; 2014;20:589–96. Available from. [CrossRef]
- Rasigade JP, Laurent F, Lina G, Meugnier H, Bes M, Vandenesch F, et al. Global distribution and evolution of panton-valentine leukocidin-positive methicillin-susceptible staphylococcus aureus, 1981-2007. Journal of Infectious Diseases. 2010;201:1589–97.
- Shittu A, Oyedara O, Abegunrin F, Okon K, Raji A, Taiwo S, et al. Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: A multicentre study in Nigeria. BMC Infect Dis. 2012;12.
- Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, Koumaré AK, et al. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J Bacteriol. 2008;190:3962–8.
- Mietze A, Morick D, Köhler H, Harrus S, Dehio C, Nolte I, et al. Combined MLST and AFLP typing of Bartonella henselae isolated from cats reveals new sequence types and suggests clonal evolution. Vet Microbiol. 2011;148:238–45.
- Jappe U, Heuck D, Strommenger B, Wendt C, Werner G, Altmann D, et al. Staphylococcus aureus in dermatology outpatients with special emphasis on community-associated methicillin-resistant strains. Journal of Investigative Dermatology. 2008;128:2655–64.
- Sudagidan M, Aydin A. Virulence properties of methicillin-susceptible Staphylococcus aureus food isolates encoding Panton-Valentine Leukocidin gene. Int J Food Microbiol [Internet]. Elsevier B.V.; 2010;138:287–91. Available from. [CrossRef]
- Rosenthal ME, Mediavilla J, Chen L, Sonnenfeld J, Pierce L, Shannon A, et al. Molecular epidemiology of Staphylococcus aureus in post-earthquake northern Haiti. International Journal of Infectious Diseases [Internet]. International Society for Infectious Diseases; 2014;29:146–51. Available from. [CrossRef]
- Fey PD, Saïd-Salim B, Rupp ME, Hinrichs SH, Boxrud DJ, Davis CC, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. American Society for Microbiology; 2003;47:196–203.
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34.
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. 2000.
- Zhu X, Sun X, Zeng Y, Feng W, Li J, Zeng J, et al. Can nasal Staphylococcus aureus screening and decolonization prior to elective total joint arthroplasty reduce surgical site and prosthesis-related infections? A systematic review and meta-analysis. J Orthop Surg Res. BioMed Central Ltd.; 2020.
- Haney Carr J, Hageman J. Guidance on the diagnosis and management of PVL-associated Staphylococcus aureus infections (PVL-SA) in England, 2 nd Edition.

| ST | spa type | isolate | AMG | BLA | FOF | ML | QA | QN | ST | TET | TMP | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aac(6')-Ie aph(2'')-Ia | aph(3')-IIIa | blaI | blaR1 | blaZ | mecA | mecI | mecR1 | fosB | glpT A100V | glpT L27F | murA E291D | murA G257D | murA T396N | erm(C) | mph(C) | msr(A) | qacC | gyrA S84L | parC S80F | parC S80Y | sat4 | tet(38) | tet(K) | dfrG | dfrS1 | |||
| 8 | t121 | 7111 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| 8 | t121 | 961 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| 8 | t1476 | 1684 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| 8 | t1476 | 2090 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 32 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 5232 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 998 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 1052 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| 8 | t1476 | 1053 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| 8 | t1476 | 1057 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 1401 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 |
| 8 | t1476 | 1405 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| 8 | t1476 | 994 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 |
| 8 | t1476 | 995 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| 8 | t1476 | 996 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| 121 | t314 | 3326 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 121 | t314 | 2777 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 121 | t314 | 4101 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 121 | t314 | 562 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 121 | t314 | 1043 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 152 | t1096 | 905 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 152 | t1096 | 907 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 152 | t355 | 2656 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| 152 | t4690 | 2007 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 152 | t4690 | 3247 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 772 | t657 | 5729 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| 789 | t091 | 1002 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| ST | spa type | isolate | Exo-enzymes | Host immunity | Toxins | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aur | splA | splB | splE | ACME | sak | scn | edinB | hlgA | hlgB | hlgC | lukD | lukE | lukF-PV | lukS-PV | sea | seb | sec | seg | sei | sej | sek | sel | sem | sen | seo | sep | seq | ser | seu | tst | |||
| 8 | t121 | 7111 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 8 | t121 | 961 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | t1476 | 1684 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 8 | t1476 | 2090 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 8 | t1476 | 32 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 8 | t1476 | 5232 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 8 | t1476 | 998 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 8 | t1476 | 1052 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 8 | t1476 | 1053 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 8 | t1476 | 1057 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 8 | t1476 | 1401 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 8 | t1476 | 1405 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | t1476 | 994 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 8 | t1476 | 995 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 8 | t1476 | 996 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0(2) | 0 | 0 |
| 121 | t314 | 3326 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| 121 | t314 | 2777 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 121 | t314 | 4101 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 121 | t314 | 562 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 121 | t314 | 1043 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 152 | t1096 | 905 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 152 | t1096 | 907 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 152 | t355 | 2656 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 152 | t4690 | 2007 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 152 | t4690 | 3247 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 772 | t657 | 5729 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0(1) | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| 789 | t091 | 1002 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





