Submitted:
05 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Subjects, sample collection and follow-up
2.2. PBMC separation
2.3. CD4+ / CD8+ T cell Immunophenotyping
2.4. Quantification of HIV-1 Viral Load
2.5. CCR5-Δ32 and CCR5-P-59029A/G Polymorphisms
2.6. Typing of HLA class I
2.7. T Cell Responses
2.7.1. HIV antigens
2.7.2. IFN-gamma Enzyme-Linked Immunospot (Elispot) Assays
2.8. Statistical Analysis
2.9. Ethics Approval
2.10. Data Availability
3. Results
3.1. Long-term non-progressors maintain CD4+ T cell counts similar to uninfected donors and TCD4/CD8+ ratio is higher in elite controllers compared to treated patients
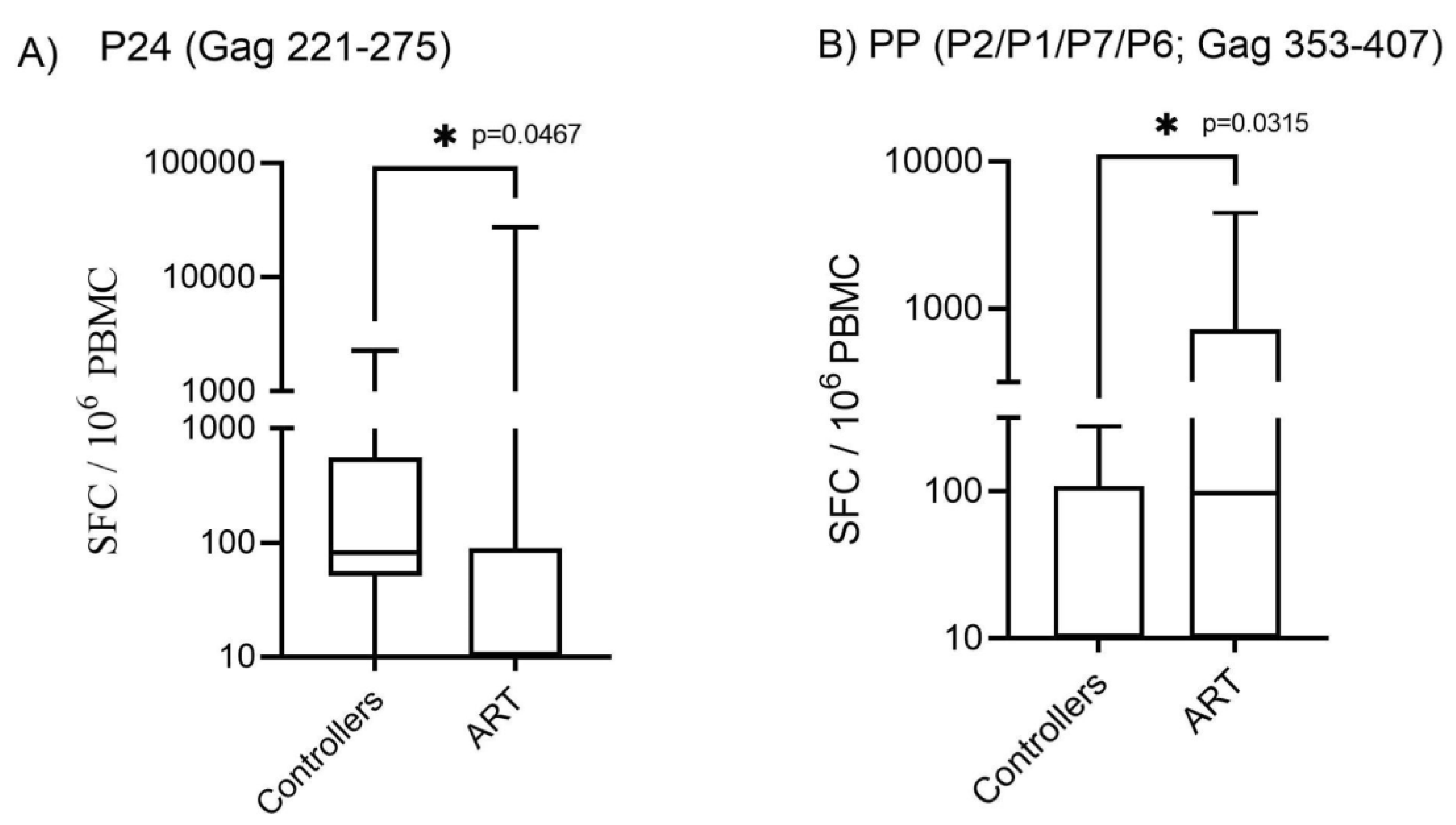
3.2. Patients with protective HLA alleles presented higher T-cell responses to HIV-1 Gag, Nef and RT pool of peptides
4. Discussion
Acknowledgments/financial disclosure
Conflicts of Interest
References
- WHO-UNAIDS – Global Aids update. Geneva 2021. Available online: https://www.unaids.org/en/resources/documents/2021/2021-global-aids-update (accessed on 18 April 2023).
- Mindel, A.; Tenant-Flowers, M. ABC of AIDS: Natural history and management of early HIV infection. BMJ 2001, 322, 1290–1293. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo-Gil, E.; Ikediobi, U.; Sutton, R.E. Mechanisms of Virologic Control and Clinical Characteristics of HIV+ Elite/Viremic Controllers. Yale J Biol Med. 2017, 90, 245–259. [Google Scholar]
- Grabar, S.; Selinger-Leneman, H.; Abgrall, S.; Pialoux, G.; Costagliola, D. Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the French Hospital Database on HIV. AIDS 2009, 23, 1163–1169. [Google Scholar] [CrossRef]
- Mandalia, S.; Westrop, S.J.; Beck, E.J.; Nelson, M.; Gazzard, B.G; Imami, N. Are long-term non-progressors very slow progressors? Insights from the Chelsea and Westminster HIV cohort, 1988-2010. PLoS One 2012, 7, e29844. [Google Scholar] [CrossRef] [PubMed]
- Gurdasani, D.; Iles, L.; Dillon, D.G.; Young, E.H.; Olson, A.D.; Naranbhai, V.; Fidler, S.; Gkrania-Klotsas, E.; Post, F.A.; Kellam, P.; Porter, K.; Sandhu, M.S.; UK HIV Genomics Consortium. A systematic review of definitions of extreme phenotypes of HIV control and progression. AIDS 2014, 28, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Salwe, S.; Padwal, V.; Velhal, S.; Sutar, J.; Bhowmick, S.; Mukherjee, S.; Nagar, V.; Patil, P.; Patel, V. Delineation of Homeostatic Immune Signatures Defining Viremic Non-progression in HIV-1 Infection. Front Immunol. 2020, 11, 182. [Google Scholar] [CrossRef]
- Graziosi, C.; Pantaleo, G.; Butini, L.; Demarest, J.F.; Saag, M.S.; Shaw, G.M.; Fauci, A.S. Kinetics of human immunodeficiency virus type 1 (HIV-1) DNA and RNA synthesis uuring Primary HIV-1 Infection. Proc Natl Acad Sci USA 1993, 90, 6405–6409. [Google Scholar] [CrossRef] [PubMed]
- Mzingwane, M.L.; Tiemessen, C.T. Mechanisms of HIV persistence in HIV reservoirs. Rev Med Virol. 2017, 27, e1924. [Google Scholar] [CrossRef]
- Virgilio, M.C.; Collins, K.L. The Impact of Cellular Proliferation on the HIV-1 Reservoir. Viruses 2020, 12, 127. [Google Scholar] [CrossRef]
- Khaitan, A.; Unutmaz, D. Revisiting immune exhaustion during HIV infection. Curr HIV/AIDS Rep. 2011, 8, 4–11. [Google Scholar] [CrossRef]
- Fenwick, C.; Joo, V.; Jacquier, P.; Noto, A.; Banga, R.; Perreau, M.; Pantaleo, G. T-cell exhaustion in HIV infection. Immunol Rev. 2019, 292, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Southern, P.J.; Reilly, C.S.; Beilman, G.J.; Chio=pman, J.G.; Schacker, T.W.; Haase, A.T. Lymphoid tissue damage in HIV-1 infection depletes naïve T cells and limits T cell reconstitution after antiretroviral therapy. PLoS Pathog. 2012, 8, e1002437. [Google Scholar] [CrossRef] [PubMed]
- Estes, J.D. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol Rev. 2013, 254, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.S.; Turner, I.; Fidler, S.; Frater, J.; Goedhals, D.; Gouder, P.; Huang, K-H.G.; Oxenius, A.; Phillips, R.; Shapiro, R.; van Vuuren, C.; McLean, A.R.; McVean, G. Mapping the drivers of within-host pathogen evolution using massive data sets. Nat Commun. 2019, 10, 3017.
- Zhao, J.; Lv, X.; Zhang, L. Ji, H. HIV-1 molecular epidemiology and drug resistance-associated mutations among treatment-naïve blood donors in China. Sci Rep. 2020, 10, 7571. [Google Scholar] [CrossRef] [PubMed]
- Magierowska, M.; Theodorou, I.; Debre, P.; Sanson, F.; Autran, B.; Riviere, Y.; Charron, D.; Franch ALT, IMMUNOCO Study Groups; Costagliola, D. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 1999, 93, 936–941.
- Saina, M.C.; Bi, X.; Lihana, R.; Lwembe, R.; Ishizaki, A.; Panikulam, A.; Palakudy, T.; Musoke, R.; Owens, M.; Songok, E.M.; Ichimura, H. Comparison of HIV-1 nef and gag Variations and Host HLA Characteristics as Determinants of Disease Progression among HIV-1 Vertically Infected Kenyan Children. PLoS One 2015, 10, e0137140. [Google Scholar] [CrossRef]
- Loureiro Dos Reis, M.M.; Queiroz, M.A.F.; Da Silva, B.C.M.; Duarte, A.J.S.; Arganaraz, G.A.; Vallinoto, A.C.R.; Arganaraz, E.R. IL6 and FAS/FASL gene polymorphisms may be associated with disease progression in HIV-1-positive ethnically mixed patients. J Med Virol. 2020, 92, 1148–1157. [Google Scholar] [CrossRef]
- Autran, B.; Descours, B.; Avettand-Fenoel, V.; Rouzioux, C. Elite controllers as a model of functional cure. Curr Opin HIV AIDS 2011, 6, 181–187. [Google Scholar] [CrossRef]
- Cockerham, L.R.; Hatano, H. Elite control of HIV: is this the right model for a functional cure? Trends Microbiol. 2015, 23, 71–75. [Google Scholar] [CrossRef]
- Lopez-Galindez, C.; Pernas, M.; Casado, C.; Olivares, I.; Lorenzo-Redondo, R. Elite controllers and lessons learned for HIV-1 cure. Curr Opin Virol. 2019, 38, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Hassaïne, G.; Agostini, I.; Candotti, D.; Bessou, G.; Caballero, M.; Agut, H.; Autran, B.; Barthalay, Y.; Vigne, R. Characterization of human immunodeficiency virus type 1 vif gene in long-term asymptomatic individuals. Virology 2000, 276, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Aquino-DeJesus, M.J.; Chan, M.; Andiman, W.A. Inhibition of human immunodeficiency virus type 1 (HIV-1) replication by a two-amino-acid insertion in HIV-1 Vif from a non progressing mother and child. J Virol. 2002, 76, 10533–10539. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Shima, T.; Nishizawa, M.; Sudo, K.; Iwamuro, S.; Okabe, T.; Takebe, Y.; Imai, M. Identification of attenuated variants of HIV-1 circulating recombinant form 01_AE that are associated with slow disease progression due to gross genetic alterations in the nef/long terminal repeat sequences. J Infect Dis. 2005, 192, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.A.; Hendel, H.; Carrington, M.; Smith, M.W.; Nelson, G.W.; O'brien, S.J.; Phair, J.; Vlahov, D.; Jacobson, L.P.; Rappaport, J.; Vasilescu, A.; Bertin-Maghit, S.; An, P.; Lu, W.; Andrieu, J.M.; Schächter, F.; Therwath, A.; Jean-François Zagury, J.F. Dominant effects of CCR2-CCR5 haplotypes in HIV-1 disease progression. J Acquir Immune Defic Syndr. 2004, 37, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- McLaren, P.J.; Coulonges, C.; Bartha, I.; Lenz, T.L.; Deutsch, A.J.; Bashirova, A.; Buchbinder, S.; Carrington, M.N.; Cossarizza, A.; Dalmau, J. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc Natl Acad Sci U S A 2015, 112, 14658–14663. [Google Scholar] [CrossRef] [PubMed]
- Fellay, J.; Ge, D.; Shianna, K.V.; Colombo, S.; Ledergerber, B.; Cirulli, E.T.; Urban, T.J.; Zhang, K.; Gumbs, C.E.; Smith, J.P. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009, 5, e1000791. [Google Scholar] [CrossRef]
- Antoni, G.; Guergnon, J.; Meaudre, C.; Samri, A.; Boufassa, F.; Goujard, C.; Lambotte, O.; Autran, B.; Rouzioux, C.; Costagliola, D.; Meyer, L. MHC-driven HIV-1 control on the long run is not systematically determined ar early times post HIV-1 infection. AIDS 2013, 27, 1707–1716. [Google Scholar] [CrossRef]
- Sundaramurthi, J.C., Ashokkumar, M., Swaminathan, S., Hanna, L.E. HLA based selection of epitopes offers a potential window of opportunity for vaccine design against HIV. Vaccine 2017, 35, 5568–5575. [CrossRef]
- Goila-Gaur, R.; Strebel, K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 2008, 5, 51. [Google Scholar] [CrossRef]
- Ruffin, N.; Brezar, V.; Ayinde, D.; Lefebvre, C.; Wiesch, J.S.Z.; van Lunzen, J.; Bockhorn, M.; Schwartz, O.; Hocini, H.; Lelievre, J-D. et al. Low SAMHD1 expression following T-cell activation and proliferation renders CD4+ T cells susceptible to HIV-1. AIDS 2015, 29, 519–530. [CrossRef] [PubMed]
- Liberatore, R.A.; Bieniasz, P.D. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci U S A. 2011, 108, 18097–18101. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.M., Malik, H.S.; Emerman, M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 2007, 316, 1756–1758. [CrossRef] [PubMed]
- Côrtes, F.H.; Bello, G.; Vorsatz, C.; Pilotto, J.H.; Guimarães, M.L.; Grinsztejn, B.; Veloso, V.G.; Pinto, A.R.; Morgado, M.G. Higher cross-subtype IFN-γ ELISpot responses to Gag and Nef peptides in Brazilian HIV-1 subtype B- and F1- than in C-infected subjects. Vaccine 2013, 31, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Côrtes, F.H.; Passaes, C.P.B.; Bello, G.; Teixeira, S.L.M.; Vorsatz, C.; Babic, D.; Sharkey, M.; Grinsztejn, B.; Veloso, V.; Stevenson, M. HIV controllers with different viral load cutoff levels have distinct virologic and immunologic profiles. J Acquir Immune Defic Syndr. 2015, 68, 377–385. [Google Scholar] [CrossRef]
- Almeida, R.R.; Rosa, D.S.; Ribeiro, S.P.; Santana, V.C.; Kallás, E.G.; Sidney, J.; Sette, A.; Kalil, J.; Cunha-Neto, E. Broad and cross-clade CD4+ T-cell responses elicited by a DNA vaccine encoding highly conserved and promiscuous HIV-1 M-group consensus peptides. PLoS One 2012, 7, e45267. [Google Scholar] [CrossRef] [PubMed]
- Laher, F.; Ranasinghe, S.; Porichis, F.; Mewalal, N.; Pretorius, K.; Ismail, N.; Buus, S.; Stryhn, A.; Carrington, M.; Walker, B.D. HIV Controllers Exhibit Enhanced Frequencies of Major Histocompatibility Complex Class II Tetramer+Gag-Specific CD4+ T Cells in Chronic Clade C HIV-1 Infection. J Virol. 2017, 91, e02477–16. [Google Scholar] [CrossRef]
- Hersperger, A.R., Migueles, S.A., Betts, M.R.; Connors, M. Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr Opin HIV AIDS 2011, 6, 169–173. [CrossRef]
- Ferre, A.L.; Lemongello, D.; Hunt, P.W.; Morris, M.M.; Garcia, J.C.; Pollard, R.B.; Yee, H.F.Jr.; Martin, J.N.; Deeks, S.G.; Shacklett, B.L. Immunodominant HIV-specific CD8+ T-cell responses are common to blood and gastrointestinal mucosa, and Gag-specific responses dominate in rectal mucosa of HIV controllers. J Virol. 2010, 84, 10354–10365. [Google Scholar] [CrossRef]
- Kristiansen, T.B.; Knudsen, T.B., Ohlendorff, S., Eugen-Olsen, J. A new multiplex PCR strategy for the simultaneous determination of four genetic polymorphisms affecting HIV-1 disease progression. J Immunol Methods 2001, 252, 147–151. [CrossRef] [PubMed]
- Dalod, M.; Dupuis, M.; Deschemin, J.C.; Goujard, C.; Deveau, C.; Meyer, L.; Ngo, N.; Rouzioux, C.; Guillet, J.G.; Delfraissy, J.F.; Sinet, M.; Venet, A. Weak anti-HIV CD8(+) T-cell effector activity in HIV primary infection. J Clin Invest. 1999, 104, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Lu, W.; Samri, A.; Costagliola, D.; Schnuriger, A.; da Silva, B.C.M.; Blanc, C.; Larsen, M.; Theodorou, I; Rouzioux, C. et al. Distinct differentiation profiles of HIV-Gag and Nef-specific central memory CD8+ T cells associated with HLA-B57/5801 and virus control. AIDS 2010, 24, 2323–2329.
- Silva, B.C.; Grassi, M.F.R.; Coutinho, R.; Mascarenhas, R.E.M.; Olavarria, V.N.; Coutinho-Borgo, A.; Kalil, J.; Cunha-Neto, E.; Fonseca, S.G. Mycobacterium tuberculosis epitope-specific interferon-g production in healthy Brazilians reactive and non-reactive to tuberculin skin test. Mem Inst Oswaldo Cruz 2014, 109, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Edwards, B.H.; Bansal, A.; Sabbaj, S.; Bakari, J.; Mulligan, M.J.; Goepfert, P.A.; Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol. 2002, 76, 2298–2305. [CrossRef] [PubMed]
- Addo, M.M.; Yu, X.G.; Rathod, A.; Cohen, D.; Eldridge, R.L.; Strick, D.; Johnston, M.N.; Cordoran, C.; Wurcel, A.G.; Fitzpatrick, C.A. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003, 77, 2081–2092. [Google Scholar] [CrossRef]
- Thakar, M.R.; Bhonge, L.S.; Lakhashe, S.K.; Shankarkumar, U.; Sane, S.S.; Kulkarni, S.S.; Mahajan, B.A.; Paranjape, R.S. Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef. J Infect Dis. 2005, 192, 749–759. [Google Scholar] [CrossRef]
- Kiepiela, P.; Ngumbela, K.; Thobakgale, C.; Ramduth, D.; Honeyborne, E.; Moodley, E.; Reddy, S.; de Pierres, C.; Mncube, Z.; Mkhwanazi, N. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007, 13, 46–53. [Google Scholar] [CrossRef]
- Zuñiga, R.; Lucchetti, A.; Galvan, P.; Sanchez, S.; Sanchez, C.; Hernandez, A.; Sanchez, H.; Frahm, N.; Linde, C.H.; Hewitt, H.S. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006, 80, 3122–3125. [Google Scholar] [CrossRef]
- Dyer, W.B.; Zaunders, J.J.; Yuan, F.F.; Wang, B.; Learmont, J.C.; Geczy, A.F.; Saksena, N.K.; McPhee, D.A.; Gorry, P.R.; Sullivan, J.S. Mechanisms of HIV non-progression; robust and sustained CD4+ T-cell proliferative responses to p24 antigen correlate with control of viraemia and lack of disease progression after long-term transfusion-acquired HIV-1 infection. Retrovirology 2008, 5, 112. [Google Scholar] [CrossRef]
- Emu, B.; Sinclair, E.; Favre, D.; Moretto, W.J.; Hsue, P.; Hoh, R.; Martin, J.N.; Nixon, D.F.; McCune, J.M.; Deeks, S.G. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005, 79, 14169–14178. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.R.; Ambrozak, D.R.; Douek, D.C.; Bonhoeffer, S.; Brenchley, J.M.; Casazza, J.P.; Koup, R.A.; Picker, L.J. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001, 75, 11983–11991. [Google Scholar] [CrossRef] [PubMed]
- Frahm, N.; Korber, B.T.; Adams, C.M.; Szinger, J.J.; Draenert, R.; Addo, M.M.; Feeney, M.E.; Yusim, K.; Sango, K.; Brown, N.V. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004, 78, 2187–2200. [Google Scholar] [CrossRef] [PubMed]
- Adland, E.; Carlson, J.M.; Paioni, P.; Kloverpris, H.; Shapiro, R.; Ogwu, A.; Riddell, L.; Luzzi, G.; Chen, F.; Balachandran, T. Nef-specific CD8+ T cell responses contribute to HIV-1 immune control. PLoS One 2013, 8, e73117. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.R.; Nason, M.C.; West, S.M.; De Rosa, S.C.; Migueles, S.A.; Abraham, J.; Lederman, M.M.; Benito, J.M.; Goepfeert, P.A.; Connors, M. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8(+) T cells. Blood 2006, 107, 4781–4789. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G.; Walker, B.D. Human immunodeficiency virus controllers: Mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 2007, 27, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Lassen, K.G.; Lobritz, M.A.; Bailey, J.R.; Johnston, S.; Nguyen, S.; Lee, B; Chou, T.; Siliciano,R.F.; Markowitz, M.; Arts, E.J. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 2009, 5, e1000377.
- Fellay, J.; Shianna, K.V.; Ge, D.; Colombo, S.; Ledergerber, B.; Weale, M.; Zhang, K.; Gumbs, C.; Castagna, A.; Cossarizza, A. A whole-genome association study of major determinants for host control of HIV-1. Science 2007, 317, 944–947. [Google Scholar] [CrossRef]
- Blankson, J.N. Effector mechanisms in HIV-1 infected elite controllers: highly active immune responses? Antiviral Res. 2010, 85, 295–302. [Google Scholar] [CrossRef]
- Lécuroux, C.; Sáez-Cirión, A.; Girault, I.; Versmisse, P.; Boufassa, F.; Avettand-Fenoël, V.; Rouzioux, C; Meyer, L.; Pancino, G.; Lambotte, O. et al. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol. 2014, 88, 176–187.
- Pereyra, F.; Addo, M.M.; Kaufmann, D.E.; Liu, Y.; Miura, T.; Rathod, A.; Baker, B.; Trocha, A.; Rosenberg, R.; Mackey, E. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008, 197, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Brener, J.; Gall, A.; Hurst, J.; Batorsky, R.; Lavandier, N.; Chen, F.; Edwards, A.; Bolton, C.; Dsouza, R.; Allen, T. Rapid HIV disease progression following superinfection in an HLA-B*27:05/B*57:01-positive transmission recipient. Retrovirology 2018, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, S.L.M.; De Sá, N.B.R.; Campos, D.P.; Coelho, A.B.; Guimarães, M.L.; Leite, T.C.N.F.; Veloso, V.G.; Morgado, M.G. Association of the HLA-B*52 allele with non-progression to AIDS in Brazilian HIV-1-infected individuals. Genes Immun. 2014, 15, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Kiepiela, P.; Leslie, A.; Honeyborne, I.; Ramduth, D.; Thobakgale, C.; Chetty, S.; Rathnavalu, P.; Moore, C.; Pfafferott, K; Hilton, L. et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004, 432, 769–775.
- de Azevedo, S.S.D.; Ribeiro-Alves, M.; Côrtes, F.H.; Delatorre, E.; Spangenberg, L.; Naya, H.; Seito, L.N.; Hoagland, B.; Grinsztejn, B.; Veloso, V.G. Increased expression of CDKN1A/p21 in HIV-1 controllers is correlated with upregulation of ZC3H12A/MCPIP1. Retrovirology 2020, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Leng, J.; Ho, H.P.; Buzon, M.J.; Pereyra, F.; Walker, B.D.; Yu, X.G.; Chang, E.J.; Lichterfeld, M. A cell-intrinsic inhibitor of HIV-1 reverse transcription in CD4(+) T cells from elite controllers. Cell Host Microbe. 2014, 15, 717–728. [Google Scholar] [CrossRef]
- Paximadis, M.; Ngqobe, R.N.; Chaisson, R.E.; Martinson, N.A.; Tiemessen, C.T. RICH2 is implicated in viraemic control of HIV-1 in black South African individuals. Infect Genet Evol. 2017, 49, 78–87. [Google Scholar] [CrossRef]
- Le Clerc, S.; Coulonges, C.; Delaneau, O.; van Manen, D.; Herbeck, J.T.; Limou, S.; An, P.; Martinson, J.J.; Spadoni, J.L.; Therwath, A. Screening low-frequency SNPS from genome-wide association study reveals a new risk allele for progression to AIDS. J Acquir Immune Defic Syndr. 2011, 56, 279–284. [Google Scholar] [CrossRef]
- Limou, S.; Coulonges, C.; Herbeck, J.T.; van Manen, D.; An, P.; Le Clerc, S.; Delaneau, O.; Diop, G.; Taing, L.; Montes, M. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term non progression to AIDS. J Infect Dis. 2010, 202, 908–915. [Google Scholar] [CrossRef]
- Stefani, C.; Sangalli, A.; Locatelli, E.; Federico, T.; Malerba, G.; Romanelli, M.G.; Argañaraz, G.A.; Da Silva, B.C.M.; Da Silva, A.J.D.; Casseb, J. Increased Prevalence of Unstable HLA-C Variants in HIV-1 Rapid-Progressor Patients. Int J Mol Sci. 2022, 23, 14852. [Google Scholar] [CrossRef]
- Luque, M.C.; Santos, C.C.; Mairena, E.C.; Wilkinson, P.; Boucher, G.; Segurado, A.C.; Fonseca, L.A.; Sabino, E.; Kalil, J.E.; Cunha-Neto, E. Gene expression profile in long-term non progressor HIV infected patients: in search of potential resistance factors. Mol Immunol. 2014, 62, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bottarel, F.; Bonissoni, S.; Lucia, M.B.; Bragardo, M.; Bensi, T.; Buonfiglio, D.; Mezzatesta, C.; DiFranco, D.; Balotta, C.; Capobianchi, M.R. Decreased function of Fas in patients displaying delayed progression of HIV-induced immune deficiency. Hematol J. 2001, 2, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Cirión, A.; Pancino, G.; Sinet, M.; Venet, M.; Lambotte, O.; ANRS EP36 HIV Controllers Study Group. HIV controllers: how do they tame the virus? Trends Immunol. 2007, 28, 532–540. [Google Scholar] [CrossRef] [PubMed]
| Variables | Controllers (C) (n = 19) |
Antiretroviral Treated Individuals (ART) (n = 18) |
P value |
|---|---|---|---|
| Age (years) [median (±SD)] | 51 (±12.08) | 56 (±10.08) | 0.6374 |
| Diagnosis (years) | 17 (±6.405) | 19.5 (±9.41) | 0.8072 |
| Nadir CD4 T cell count (x 106/L) [median (±SD)] | 596 (±186) | 315 (±201) | <0.001 |
| CD4 T cell count (x 106/L) [median (±SD)] | 850 (±331) | 637 (±317) | 0.0073 |
| CD8 T cell count (x 106/L) [median (±SD)] | 978 (±612) | 1104 (±445) | 0.5655 |
| Log10 plasma VL (copies/mL) [median (±SD)] | 704 (±3702) | 25 (±23704) | 0.2239 |
| Peptides | HLA | |||||
|---|---|---|---|---|---|---|
| Locus A1 | Locus A2 | Locus B1 | Locus B2 | Locus C1 | Locus C2 | |
|
P17-1 (Gag 1-55) |
.515 | .077 | .623 | .308 | .620 | .236 |
|
P17-2 (Gag 45-99) |
.523 | .374 | .834 | .104 | .032 | .568 |
|
P17-3 (Gag 89-143) |
.880 | .738 | .304 | .847 | .242 | .774 |
|
P24-1 (Gag 133-187) |
.601 | .602 | .510 | .398 | .726 | .855 |
|
P24-2 (Gag 177-231) |
.231 | .896 | .305 | .152 | .841 | .247 |
|
P24-3 (Gag 221-275) |
.508 | .512 | .175 | .462 | .021 | .459 |
|
P24-4 (Gag 265-319) |
.494 | .413 | .273 | .579 | .181 | .533 |
|
P24-5 (Gag 309-363) |
.532 | .434 | .159 | .906 | .745 | .375 |
|
P2/P1/P7/P6 1 (Gag 353-407) |
.067 | .454 | .009 | .221 | .578 | .127 |
|
P2/P1/P7/P6 2 (Gag 397-451) |
.319 | .572 | .444 | .202 | .801 | .536 |
|
P2/P1/P7/P6 3 (Gag 441-500) |
.291 | .586 | .429 | .549 | .979 | .257 |
|
Nef1 (Nef 1-55) |
.844 | .312 | .145 | .791 | .537 | .071 |
|
Nef2 (Nef 45-103) |
.654 | .507 | .088 | .415 | .863 | .110 |
|
Nef3 (Nef 93-155) |
.093 | .637 | .033 | .094 | .080 | .062 |
|
Nef4 (Nef 145-206) |
.026 | .686 | .002 | .070 | .932 | .053 |
|
RT1 (RT 153-223) |
.070 | .055 | .095 | .590 | .380 | .431 |
|
RT2 (RT 213-283) |
.442 | .880 | .012 | .063 | .770 | .712 |
|
RT3 (RT 273-343) |
.449 | .771 | .065 | .786 | .761 | .372 |
|
RT4 (RT 333-395) |
.134 | .249 | .395 | .027 | .666 | .323 |
|
RT5 (RT 385-455) |
.224 | .944 | .299 | .485 | .851 | .510 |
|
RT6 (RT 445-503) |
.630 | .537 | .347 | .810 | .626 | .280 |
|
RT7 (RT 493-551) |
.714 | .521 | .127 | .390 | .567 | .991 |
|
RT8 (RT 541-599) |
.207 | .805 | .510 | .023 | .975 | .919 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





