Submitted:
29 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.2. Kidney top expressed miRNAs and kidney enriched miRNAs in uEV
2.3. Literature review of miRNAs associated with DKD
2.4. Stable mRNAs across datasets
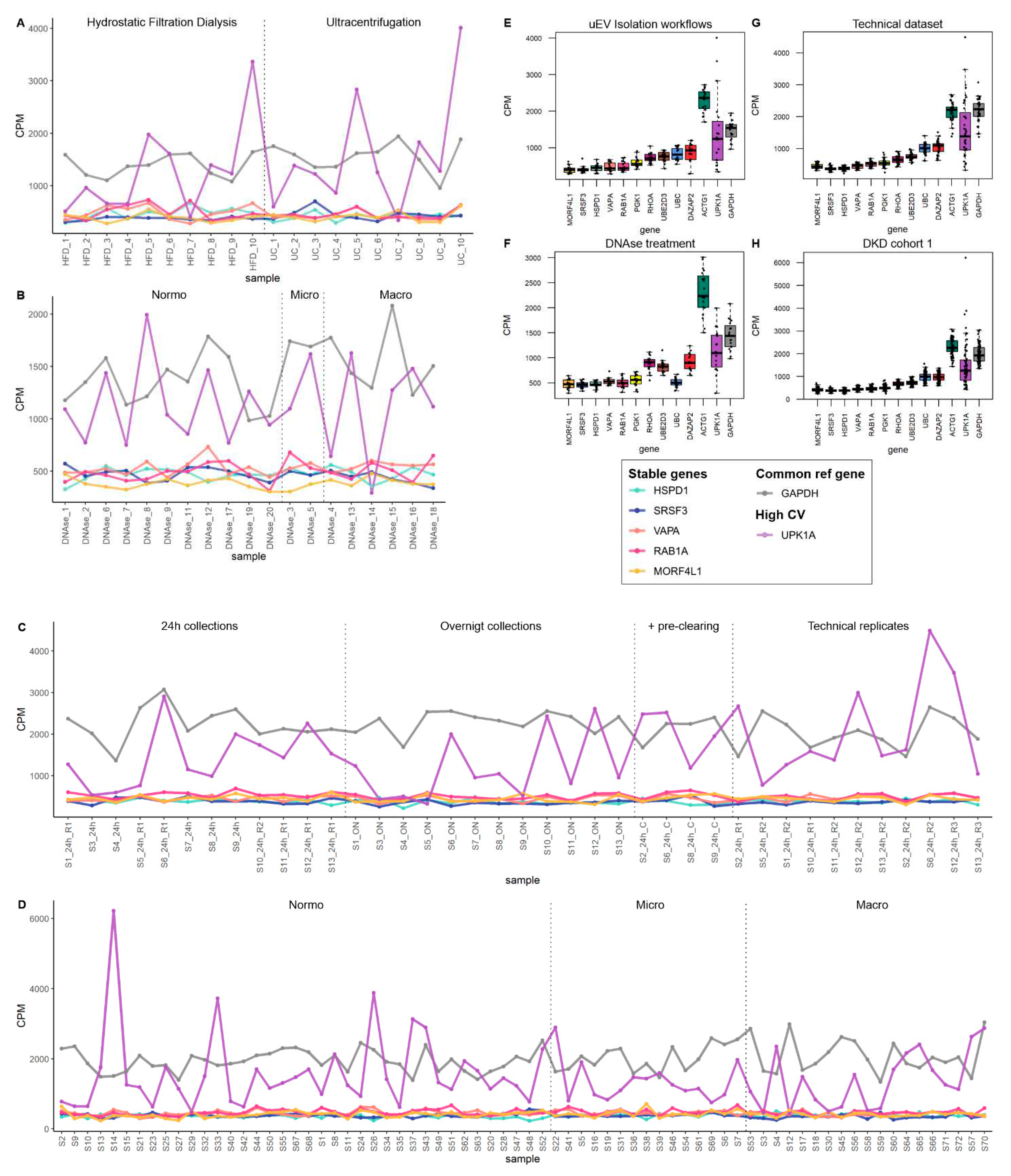
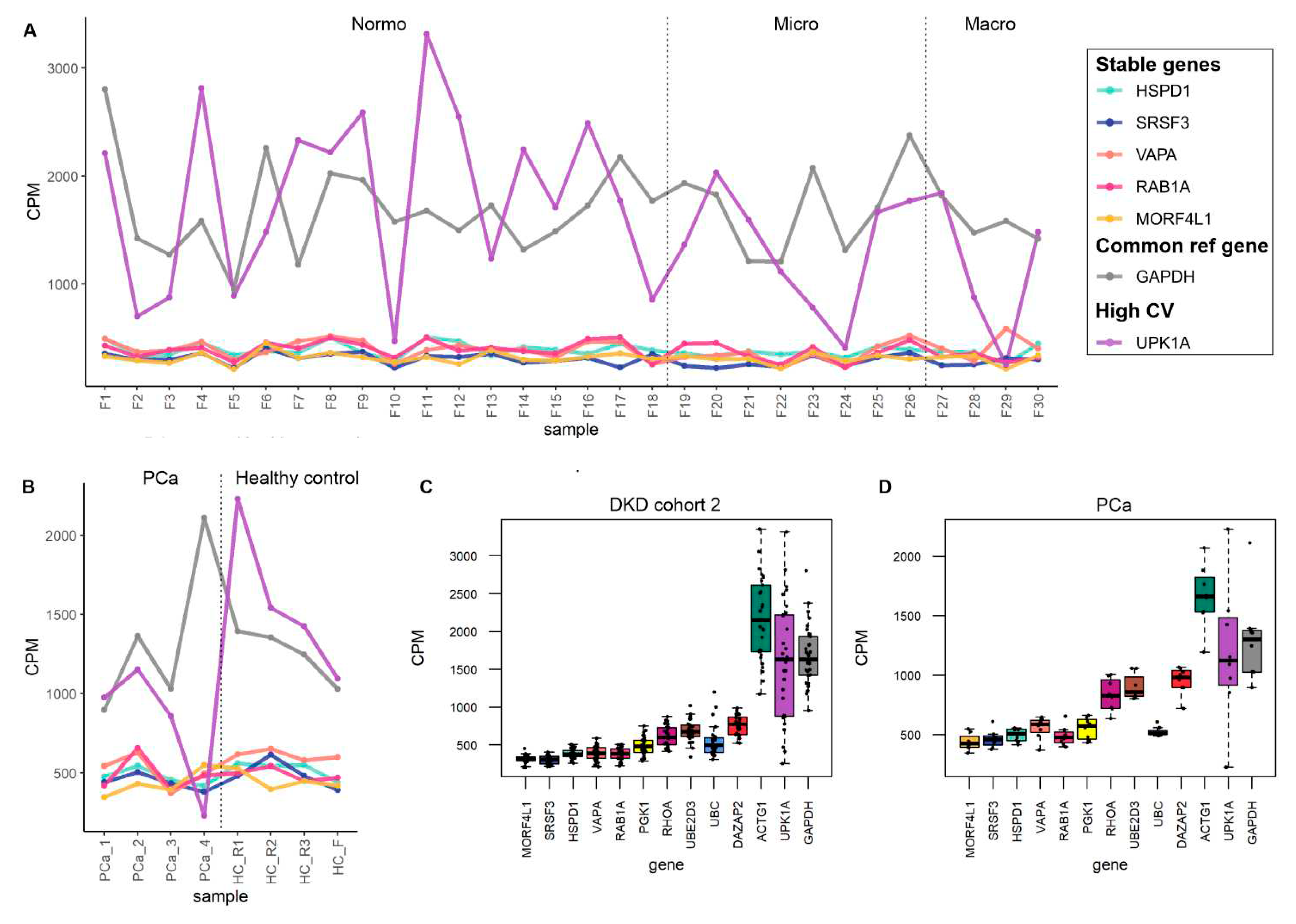
2.5. Data visualization
3. Results
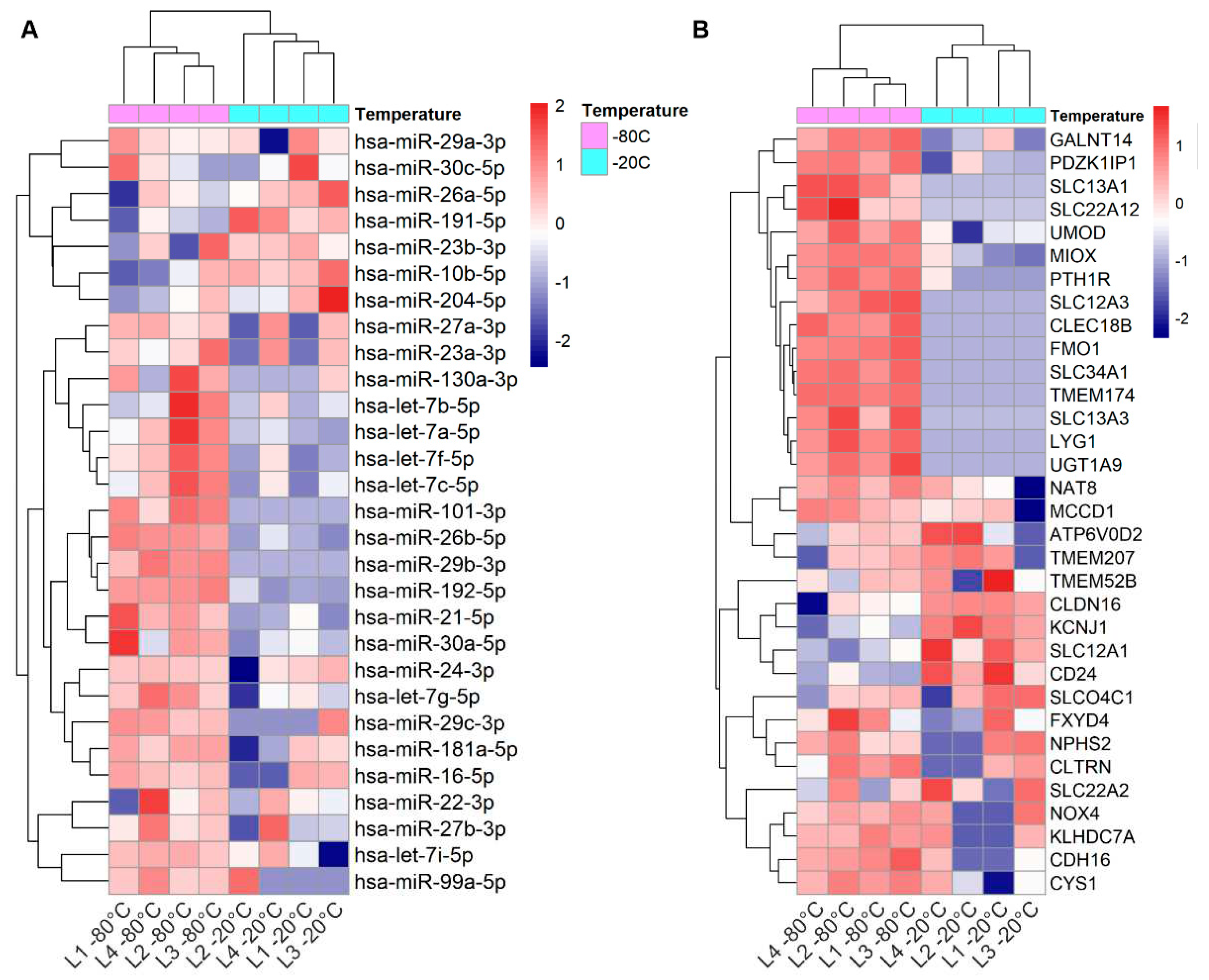
3.1. Dysregulated miRNAs in samples stored at suboptimal temperature: significance for kidney disease biomarker discovery
3.2. Effect of pre-analytical variables on kidney transcriptome in uEV isolates
3.2.1. Effect of storage temperature
3.2.2. Effect of isolation workflows
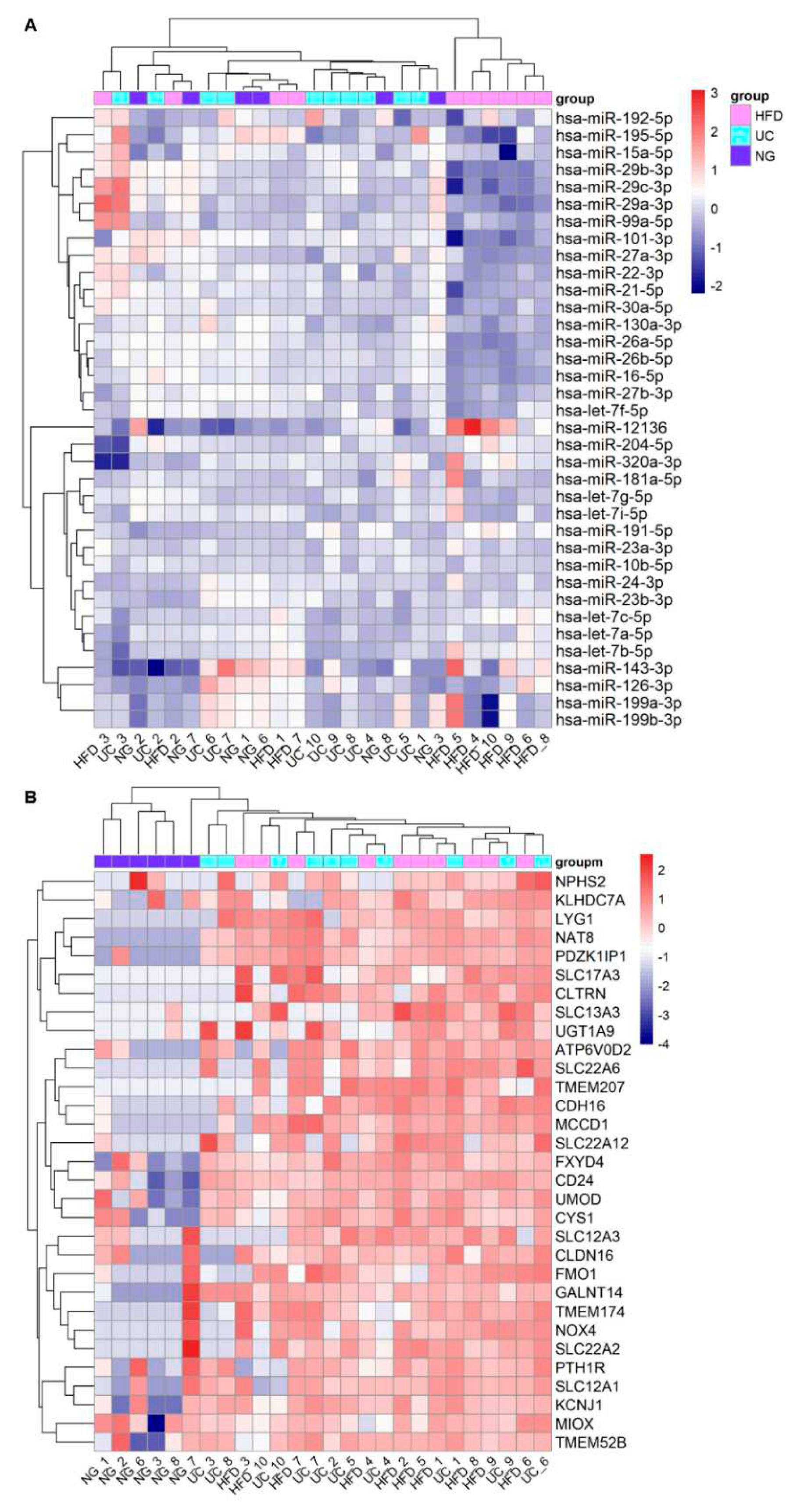
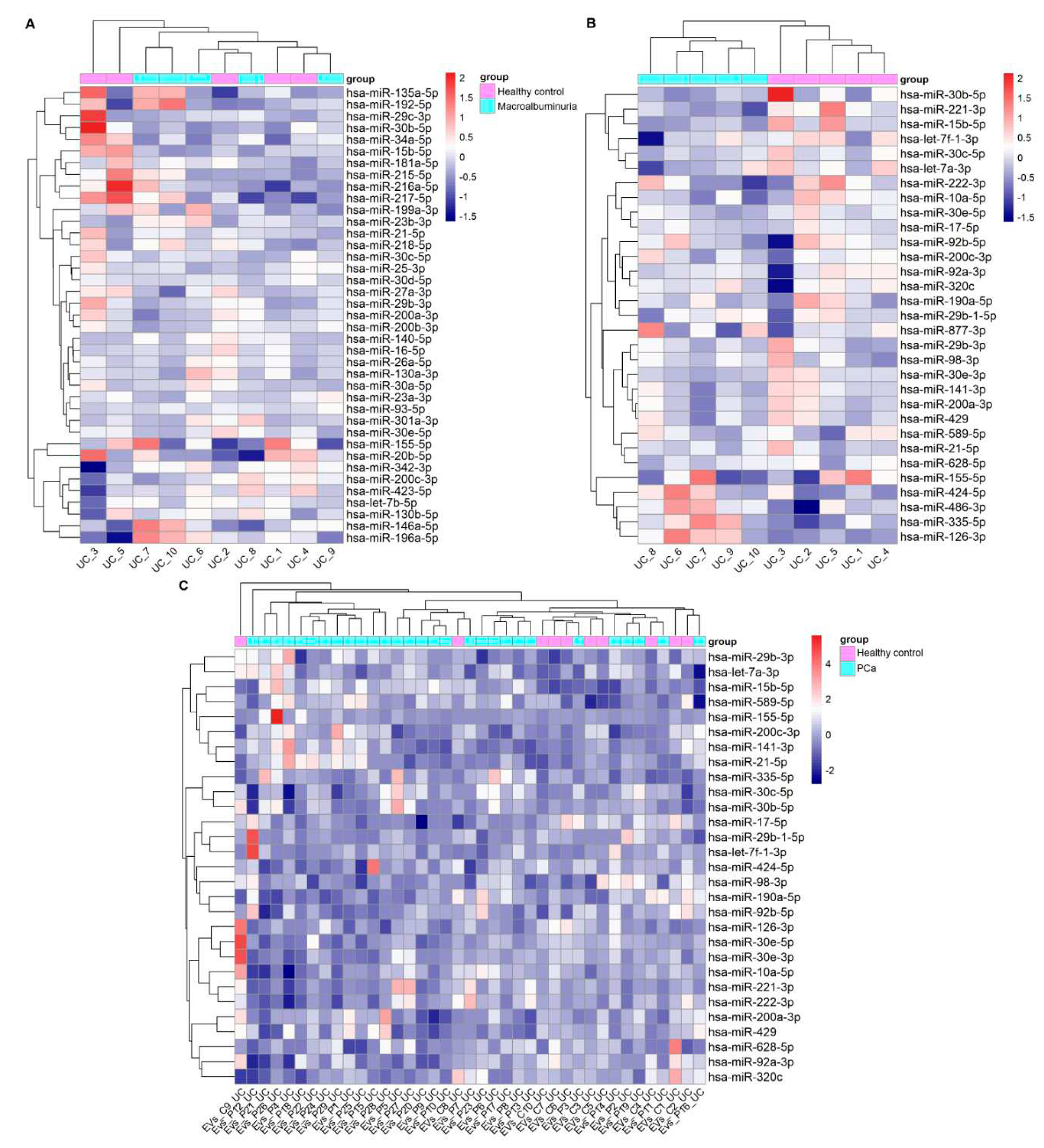
3.3. Replication of DKD –associated miRNA by UC –based uEV isolation and sequencing workflow
| Sample | Groups | Upregulated miRNAs | Downregulated miRNAs | Reference |
|---|---|---|---|---|
| Urine | Urine from T1D (Normal, overt nephropathy, intermittent microalbuminuria, persistent microalbuminuria) | DKD vs non DKD: miR-619, miR-486-3p, miR-335-5p, miR-552, miR-1912, miR-1124-3p, miR-424-5p, miR-141-3p, miR-29b-1-5p | DKD vs non-DKD: miR-221-3p | (Argyropoulos et al. 2013) |
| MA vs baseline: miR-214-3p, miR-92b-5p, miR-765, miR-429, miR-373-5p, miR-1913, miR-638 | MA vs baseline: miR-323b-5p, miR-221-3p, miR-524-5p, miR-188-3p | |||
| PMA vs IMA: miR323b-5p, miR-433, miR-17-5p, miR-222-3p, 628-5p | PMA vs IMA: miR-589-5p, miR-373-5p, miR-92a-3p | |||
| Urinary sediments | Diabetic glomerulosclerosis, minimal change nephropathy or focal glomerulosclerosis, membranous nephropathy, and healthy donors | miR-200c | miR-638, miR-192 | (Wang et al. 2013) |
| uEV** | T1D with normoalbuminuria and microalbuminuria and non-diabetic controls | miR-130a, miR-145 | miR-155, miR-424 | (Barutta et al. 2013) |
| Urine | T2D DKD, T2D, and healthy donors | miR-126 (T2D DKD > T2D) | (Liu et al. 2014) | |
| uEV | T2D normoalbuminuric, microalbuminuric, or macroalbuminuric | microalbuminuria vs normoalbuminuria and controls: miR-192, miR-194, and miR-215. | macroalbuminuria vs microalbuminuria: miR-192, miR215 | (Jia et al. 2016) |
| uEV* | T2D DKD, T2D, and healthy donors | miR-320c, miR-6068 | (Delić et al. 2016) | |
| urine pellets and uEV * | T2D albuminuric, normoalbuminuric, and healthy controls | miR-15b, miR-34a, miR-636 | (Eissa et al. 2016) | |
| uEV* | T2D normoalbuminuria and microalbuminuria | miR-877-3p | (Xie et al. 2017) | |
| uEV** | T1D normoalbuminuria, intermittent macroalbuminuria, persistent macroalbuminuria, and overt macroalbuminuria | Overt vs normal: miR-26a-1-5p, miR-30-5p PMA vs IMA/ non microalbuminuria: miR-200c-3p | Overt vs normal: miR144-3p | (Ghai et al. 2018) |
| Urine** | PMA vs IMA: miR-10a-5p, miR-200a-3p | |||
| Urine* | Diabetic, DKD and healthy donors | miR-126-3p, miR-155-5p, and miR-29b-3p | (Beltrami et al. 2018) | |
| Urine* | Urine and plasma from T1D and DKD | miR-30e-5p | (Dieter et al. 2019) | |
| uEV* | T2D DKD, T2D normal renal function, and non-T2D CKD | miR-21-5p | miR-30b-5p | (Zang et al. 2019) |
| Urine** | DKD and non-diabetic renal disease | T2D vs non-diabetic renal disease: miR-27-3p, miR-1228 | (Conserva et al. 2019) | |
| uEV* | T2D and normoalbuminuria, microalbuminuria or macroalbuminuria and healthy donors | miR-15b-5p | (Tsai et al. 2020) | |
| uEV* | TD2 DKD and healthy donors | miR-30e-3p, miR-30c-5p, miR-190a-5p, miR-98-3p, let-7a-3p, miR-30b-5p, and let-7f-1-3p | (Park et al. 2022) |
| Regulation | ||||
| DKD in UC dataset |
uEV/ urine/ Urine sediments literature |
Examples of association with diabetic Kidney disease or kidney diseases, or pathways associated with DKD (e.g. fibrosis, inflammation, autophagy, and oxidative stress) | ||
| Cluster 1 | miR-30b-5p | down | down | In hyperglycemic conditions, expression levels reduced in HK-2 cells and epithelial-mesenchyma increased (Wang et al. 2021) |
| miR-221-3p | down | down | Associated with tumorigenesis and prognosis of kidney renal clear cell carcinoma (Zhou et al. 2022). In HUVEC cells, hyperglycemia induced this miRNA and was associated with impairment of endothelial cell migration and homing mediated (Li et al. 2009). | |
| miR-15b-5p | down | up | Up regulated in urine from db/db mouse and T2D patients. In mesangial cell lines hyperglycemia upregulates this miRNA and targets BCL-2 inducing apoptosis (Tsai et al. 2020). | |
| let-7f-1-3p | down | down | Upregulated in the kidney of a rat model of AKI (Liu et al. 2019). By regulating negatively STAT3, induces autophagy in glioma cells (Yang et al. 2020). | |
| miR-30c-5p | down | down | Downregulate in kidney tissue from diabetic patients and high glucose-induced HK-2 cells. Downregulation causes epithelial to mesenchymal transition mediated by ROCK2 (Cui, Yu, and Cui 2020). | |
| Cluster 4 | miR-424-5p | up | up | Upregulated in high fat diet induced mouse and hepatocytes treated with palmitate. miR-424-5p suppressed insulin receptor expression in hepatocytes i.e. role in insulin resistance(Min, Yang, and Lee 2018). |
| miR-486-3p | up | up | Downregulated in biopsies from patients with diabetic nephropathy (Baker et al. 2017) | |
| miR-335-5p | up | up | In childhood asthma, the upregulation of this miRNA is associated with reduced inflammation, fibrosis and autophagy by regulating ATG5 (Liang et al. 2022). | |
| miR-126-3p | up | up | Increased in kidney biopsies from patients with DKD (Beltrami et al. 2018) | |
3.4. Exploratory analysis of reference mRNA in uEV
4. Discussion
Supplementary Materials
Authors’ Contributions
Financial Support and Sponsorship
Availability of Data and Materials
Acknowledgments
Conflicts of Interest
Declarations
References
- Abbas-Aghababazadeh, F.; Li, Q.; Fridley, B.L. Comparison of normalization approaches for gene expression studies completed with high-throughput sequencing. PLOS ONE 2018, 13, e0206312. [Google Scholar] [CrossRef]
- Agudiez, M.; Martinez, P.J.; Martin-Lorenzo, M.; Heredero, A.; Santiago-Hernandez, A.; Molero, D.; Garcia-Segura, J.M.; Aldamiz-Echevarria, G.; Alvarez-Llamas, G. Analysis of urinary exosomal metabolites identifies cardiovascular risk signatures with added value to urine analysis. BMC Biol. 2020, 18, 1–9. [Google Scholar] [CrossRef]
- Alvarez, M. L. M. Khosroheidari, E. Eddy, and J. Kiefer. 2013. ’Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy’, PLoS One, 8: e77468.
- Anders, S. , and W. Huber. 2010. ’Differential expression analysis for sequence count data’, Genome Biol, 11: R106.
- Argyropoulos, C. K. Wang, S. McClarty, D. Huang, J. Bernardo, D. Ellis, T. Orchard, D. Galas, and J. Johnson. 2013. ’Urinary microRNA profiling in the nephropathy of type 1 diabetes’, PLoS One, 8: e54662.
- Armstrong, D.A.; Dessaint, J.A.; Ringelberg, C.S.; Hazlett, H.F.; Howard, L.; Abdalla, M.A.; Barnaby, R.L.; Stanton, B.A.; Cervinski, M.A.; Ashare, A. Pre-Analytical Handling Conditions and Small RNA Recovery from Urine for miRNA Profiling. J. Mol. Diagn. 2018, 20, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Geng, J.; Zhou, Z.; Tian, J.; Li, X. MicroRNA-130b improves renal tubulointerstitial fibrosis via repression of Snail-induced epithelial-mesenchymal transition in diabetic nephropathy. Sci. Rep. 2016, 6, 20475. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.A.; Davis, S.J.; Liu, P.; Pan, X.; Williams, A.M.; Iczkowski, K.A.; Gallagher, S.T.; Bishop, K.; Regner, K.R.; Liu, Y.; et al. Tissue-Specific MicroRNA Expression Patterns in Four Types of Kidney Disease. J. Am. Soc. Nephrol. 2017, 28, 2985–2992. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, K.; Dwivedi, O.P.; Leparc, G.; Rolser, M.; Delic, D.; Forsblom, C.; Groop, P.; Groop, L.; Huber, T.B.; Puhka, M.; et al. Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. J. Extracell. Vesicles 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, K. O. P. Dwivedi, S. Valkonen, P. H. Groop, T. Tuomi, H. Holthofer, A. Rannikko, M. Yliperttula, P. Siljander, S. Laitinen, E. Serkkola, T. Af Hällström, C. Forsblom, L. Groop, and M. Puhka. 2021. ’Urinary extracellular vesicles: Assessment of pre-analytical variables and development of a quality control with focus on transcriptomic biomarker research’, J Extracell Vesicles, 10: e12158.
- Barutta, F.; Tricarico, M.; Corbelli, A.; Annaratone, L.; Pinach, S.; Grimaldi, S.; Bruno, G.; Cimino, D.; Taverna, D.; Deregibus, M.C.; et al. Urinary Exosomal MicroRNAs in Incipient Diabetic Nephropathy. PLOS ONE 2013, 8, e73798. [Google Scholar] [CrossRef] [PubMed]
- Bazzell, B.G.; Rainey, W.E.; Auchus, R.J.; Zocco, D.; Bruttini, M.; Hummel, S.L.; Byrd, J.B. Human Urinary mRNA as a Biomarker of Cardiovascular Disease. Circ. Genom. Precis. Med. 2018, 11, e002213–e002213. [Google Scholar] [CrossRef]
- Beltrami, C. K. Simpson, M. Jesky, A. Wonnacott, C. Carrington, P. Holmans, L. Newbury, R. Jenkins, T. Ashdown, C. Dayan, S. Satchell, P. Corish, P. Cockwell, D. Fraser, and T. Bowen. 2018. ’Association of Elevated Urinary miR-126, miR-155, and miR-29b with Diabetic Kidney Disease’, Am J Pathol, 188: 1982-92.
- Bera, A.; Das, F.; Ghosh-Choudhury, N.; Mariappan, M.M.; Kasinath, B.S.; Choudhury, G.G. Reciprocal regulation of miR-214 and PTEN by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. Am. J. Physiol. Physiol. 2017, 313, C430–C447. [Google Scholar] [CrossRef]
- Bhatt, K.; Lanting, L.L.; Jia, Y.; Yadav, S.; Reddy, M.A.; Magilnick, N.; Boldin, M.; Natarajan, R. Anti-Inflammatory Role of MicroRNA-146a in the Pathogenesis of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2016, 27, 2277–2288. [Google Scholar] [CrossRef]
- Bijkerk, R. R. G. de Bruin, C. van Solingen, J. M. van Gils, J. M. Duijs, E. P. van der Veer, T. J. Rabelink, B. D. Humphreys, and A. J. van Zonneveld. 2016. ’Silencing of microRNA-132 reduces renal fibrosis by selectively inhibiting myofibroblast proliferation’, Kidney Int, 89: 1268-80.
- Blijdorp, C.J.; Hartjes, T.A.; Wei, K.; van Heugten, M.H.; Bovée, D.M.; Budde, R.P.; van de Wetering, J.; Hoenderop, J.G.; van Royen, M.E.; Zietse, R.; et al. Nephron mass determines the excretion rate of urinary extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12181. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Zhong, X.; Huang, X.R.; Meng, X.-M.; You, Y.; Chung, A.C.; Lan, H.Y. MicroRNA-29b Inhibits Diabetic Nephropathy in db/db Mice. Mol. Ther. 2014, 22, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, L.; Xing, Y.; Lin, B. RETRACTED: Down-regulation of microRNA-21 reduces inflammation and podocyte apoptosis in diabetic nephropathy by relieving the repression of TIMP3 expression. Biomed. Pharmacother. 2018, 108, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Wang, X.X.; Yao, X.M.; Zhang, D.L.; Yang, X.F.; Tian, S.F.; Wang, N.S. Abated microRNA-195 expression protected mesangial cells from apoptosis in early diabetic renal injury in mice. J. Nephrol. 2011, 25, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Chorley, B. N. H. Ellinger-Ziegelbauer, M. Tackett, F. J. Simutis, A. H. Harrill, J. McDuffie, E. Atabakhsh, R. Nassirpour, L. O. Whiteley, J. F. Léonard, G. K. Carswell, E. Harpur, C. L. Chen, and J. C. Gautier. 2021. ’Urinary miRNA Biomarkers of Drug-Induced Kidney Injury and Their Site Specificity Within the Nephron’, Toxicol Sci, 180: 1-16.
- Conserva, F.; Barozzino, M.; Pesce, F.; Divella, C.; Oranger, A.; Papale, M.; Sallustio, F.; Simone, S.; Laviola, L.; Giorgino, F.; et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of Kidney Fibrosis in Diabetic Nephropathy. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cui, L. M. Yu, and X. Cui. 2020. ’MiR-30c-5p/ROCK2 axis regulates cell proliferation, apoptosis and EMT via the PI3K/AKT signaling pathway in HG-induced HK-2 cells’, Open Life Sci, 15: 959-70.
- Dai, Y.; Cao, Y.; Köhler, J.; Lu, A.; Xu, S.; Wang, H. Unbiased RNA-Seq-driven identification and validation of reference genes for quantitative RT-PCR analyses of pooled cancer exosomes. BMC Genom. 2021, 22, 1–13. [Google Scholar] [CrossRef]
- De Freitas, R.C.C.; Hirata, R.D.C.; Hirata, M.H.; Aikawa, E. Circulating Extracellular Vesicles As Biomarkers and Drug Delivery Vehicles in Cardiovascular Diseases. Biomolecules 2021, 11, 388. [Google Scholar] [CrossRef]
- DeFronzo, R.A.; Reeves, W.B.; Awad, A.S. Pathophysiology of diabetic kidney disease: impact of SGLT2 inhibitors. Nat. Rev. Nephrol. 2021, 17, 319–334. [Google Scholar] [CrossRef]
- Delić, D.; Eisele, C.; Schmid, R.; Baum, P.; Wiech, F.; Gerl, M.; Zimdahl, H.; Pullen, S.S.; Urquhart, R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLOS ONE 2016, 11, e0150154. [Google Scholar] [CrossRef]
- Deshpande, S.D.; Putta, S.; Wang, M.; Lai, J.Y.; Bitzer, M.; Nelson, R.G.; Lanting, L.L.; Kato, M.; Natarajan, R. Transforming Growth Factor-β–Induced Cross Talk Between p53 and a MicroRNA in the Pathogenesis of Diabetic Nephropathy. Diabetes 2013, 62, 3151–3162. [Google Scholar] [CrossRef] [PubMed]
- Dey, N. F. Das, M. M. Mariappan, C. C. Mandal, N. Ghosh-Choudhury, B. S. Kasinath, and G. G. Choudhury. 2011. ’MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes’, J Biol Chem, 286: 25586-603.
- Dieter, C.; Assmann, T.S.; Costa, A.R.; Canani, L.H.; de Souza, B.M.; Bauer, A.C.; Crispim, D. MiR-30e-5p and MiR-15a-5p Expressions in Plasma and Urine of Type 1 Diabetic Patients With Diabetic Kidney Disease. Front. Genet. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Dong, L. R. C. Zieren, K. Horie, C. J. Kim, E. Mallick, Y. Jing, M. Feng, M. D. Kuczler, J. Green, S. R. Amend, K. W. Witwer, T. M. de Reijke, Y. K. Cho, K. J. Pienta, and W. Xue. 2020. ’Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium’, J Extracell Vesicles, 10: e12044.
- Duan, Y.; Chen, B.; Chen, F.; Yang, S.; Zhu, C.; Ma, Y.; Li, Y.; Shi, J. Exosomal microRNA-16-5p from human urine-derived stem cells ameliorates diabetic nephropathy through protection of podocyte. J. Cell. Mol. Med. 2019, 25, 10798–10813. [Google Scholar] [CrossRef]
- Dwivedi, O.P.; Barreiro, K.; Käräjämäki, A.; Valo, E.; Giri, A.K.; Prasad, R.B.; Das Roy, R.; Thorn, L.M.; Rannikko, A.; Holthöfer, H.; et al. Genome-wide mRNA profiling in urinary extracellular vesicles reveals stress gene signature for diabetic kidney disease. iScience 2023, 26, 106686. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Aboushahba, R.; Bekhet, M.M.; Soliman, Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J. Diabetes its Complicat. 2016, 30, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Erdbrugger, U. and T. H. Le. 2016. ’Extracellular Vesicles in Renal Diseases: More than Novel Biomarkers?’, J Am Soc Nephrol, 27: 12-26.
- Erdbrügger, U. C. J. Blijdorp, I. V. Bijnsdorp, F. E. Borràs, D. Burger, B. Bussolati, J. B. Byrd, A. Clayton, J. W. Dear, J. M. Falcón-Pérez, C. Grange, A. F. Hill, H. Holthöfer, E. J. Hoorn, G. Jenster, C. R. Jimenez, K. Junker, J. Klein, M. A. Knepper, E. H. Koritzinsky, J. M. Luther, M. Lenassi, J. Leivo, I. Mertens, L. Musante, E. Oeyen, M. Puhka, M. E. van Royen, C. Sánchez, C. Soekmadji, V. Thongboonkerd, V. van Steijn, G. Verhaegh, J. P. Webber, K. Witwer, P. S. T. Yuen, L. Zheng, A. Llorente, and E. S. Martens-Uzunova. 2021. ’Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles’, J Extracell Vesicles, 10: e12093.
- Fu, Y.; Zhang, Y.; Wang, Z.; Wang, L.; Wei, X.; Zhang, B.; Wen, Z.; Fang, H.; Pang, Q.; Yi, F. Regulation of NADPH Oxidase Activity Is Associated with miRNA-25-Mediated NOX4 Expression in Experimental Diabetic Nephropathy. Am. J. Nephrol. 2010, 32, 581–589. [Google Scholar] [CrossRef]
- Gao, Y. W. Xu, C. Guo, and T. Huang. 2022. ’GATA1 regulates the microRNA-328-3p/PIM1 axis via circular RNA ITGB1 to promote renal ischemia/reperfusion injury in HK-2 cells’, Int J Mol Med, 50.
- García-Flores, M. C. M. Sánchez-López, M. Ramírez-Calvo, A. Fernández-Serra, A. Marcilla, and J. A. López-Guerrero. 2021. ’Isolation and characterization of urine microvesicles from prostate cancer patients: different approaches, different visions’, BMC Urol, 21: 137.
- Ghai, V.; Wu, X.; Bheda-Malge, A.; Argyropoulos, C.P.; Bernardo, J.F.; Orchard, T.; Galas, D.; Wang, K. Genome-wide Profiling of Urinary Extracellular Vesicle microRNAs Associated With Diabetic Nephropathy in Type 1 Diabetes. Kidney Int. Rep. 2017, 3, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Gorji-Bahri, G. N. Moradtabrizi, F. Vakhshiteh, and A. Hashemi. 2021. ’Validation of common reference genes stability in exosomal mRNA-isolated from liver and breast cancer cell lines’, Cell Biol Int, 45: 1098-110.
- Gouin, K. K. Peck, T. Antes, J. L. Johnson, C. Li, S. D. Vaturi, R. Middleton, G. de Couto, A. S. Walravens, L. Rodriguez-Borlado, R. R. Smith, L. Marbán, E. Marbán, and A. G. Ibrahim. 2017. ’A comprehensive method for identification of suitable reference genes in extracellular vesicles’, J Extracell Vesicles, 6: 1347019.
- Habuka, M. L. Fagerberg, B. M. Hallström, C. Kampf, K. Edlund, Å Sivertsson, T. Yamamoto, F. Pontén, M. Uhlén, and J. Odeberg. 2014. ’The kidney transcriptome and proteome defined by transcriptomics and antibody-based profiling’, PLoS One, 9: e116125.
- Han, X.; Li, Q.; Wang, C.; Li, Y. MicroRNA-204-3p Attenuates High Glucose-Induced MPC5 Podocytes Apoptosis by Targeting Braykinin B2 Receptor. Exp. Clin. Endocrinol. Diabetes 2018, 127, 387–395. [Google Scholar] [CrossRef]
- He, F.; Peng, F.; Xia, X.; Zhao, C.; Luo, Q.; Guan, W.; Li, Z.; Yu, X.; Huang, F. MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia 2014, 57, 1726–1736. [Google Scholar] [CrossRef]
- He, X.; Zeng, X. LncRNA SNHG16 Aggravates High Glucose-Induced Podocytes Injury in Diabetic Nephropathy Through Targeting miR-106a and Thereby Up-Regulating KLF9. Diabetes, Metab. Syndr. Obesity: Targets Ther. 2020, 13, 3551–3560. [Google Scholar] [CrossRef]
- Hogan, M.C.; Lieske, J.C.; Lienczewski, C.C.; Nesbitt, L.L.; Wickman, L.T.; Heyer, C.M.; Harris, P.C.; Ward, C.J.; Sundsbak, J.L.; Manganelli, L.; et al. Strategy and rationale for urine collection protocols employed in the NEPTUNE study. BMC Nephrol. 2015, 16, 190. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y. Y. Li, Y. Wang, W. Li, and Z. Xiao. 2021. ’Screening and Analysis of Key Genes in miRNA-mRNA Regulatory Network of Membranous Nephropathy’, J Healthc Eng, 2021: 5331948.
- Hsu, Y.-C.; Chang, P.-J.; Ho, C.; Huang, Y.-T.; Shih, Y.-H.; Wang, C.-J.; Lin, C.-L. Protective effects of miR-29a on diabetic glomerular dysfunction by modulation of DKK1/Wnt/β-catenin signaling. Sci. Rep. 2016, 6, 30575. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Li, L.; Su, B.; Yang, L.; Fan, W.; Yin, Q.; Chen, L.; Cui, T.; Zhang, J.; et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC Nephrol. 2014, 15, 142–142. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Guan, M.; Zheng, Z.; Zhang, Q.; Tang, C.; Xu, W.; Xiao, Z.; Wang, L.; Xue, Y. miRNAs in Urine Extracellular Vesicles as Predictors of Early-Stage Diabetic Nephropathy. J. Diabetes Res. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Jiang, Y. W. Wang, Z. Y. Liu, Y. Xie, Y. Qian, and X. N. Cai. 2018. ’Overexpression of miR-130a-3p/301a-3p attenuates high glucose-induced MPC5 podocyte dysfunction through suppression of TNF-α signaling’, Exp Ther Med, 15: 1021-28.
- Jiang, Z.; Tang, Y.; Song, H.; Yang, M.; Li, B.; Ni, C. miRNA-342 suppresses renal interstitial fibrosis in diabetic nephropathy by targeting SOX6. Int. J. Mol. Med. 2019, 45, 45–52. [Google Scholar] [CrossRef]
- Jones, T.F.; Bekele, S.; O’dwyer, M.J.; Prowle, J.R. MicroRNAs in Acute Kidney Injury. Nephron 2018, 140, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Arce, L.; Wang, M.; Putta, S.; Lanting, L.; Natarajan, R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011, 80, 358–368. [Google Scholar] [CrossRef]
- Kato, M. S. Putta, M. Wang, H. Yuan, L. Lanting, I. Nair, A. Gunn, Y. Nakagawa, H. Shimano, I. Todorov, J. J. Rossi, and R. Natarajan. 2009. ’TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN’, Nat Cell Biol, 11: 881-9.
- Kato, M. L. Wang, S. Putta, M. Wang, H. Yuan, G. Sun, L. Lanting, I. Todorov, J. J. Rossi, and R. Natarajan. 2010. ’Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells’, J Biol Chem, 285: 34004-15.
- Kato, M.; Wang, M.; Chen, Z.; Bhatt, K.; Oh, H.J.; Lanting, L.; Deshpande, S.; Jia, Y.; Lai, J.Y.; O’connor, C.L.; et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat. Commun. 2016, 7, 12864. [Google Scholar] [CrossRef]
- Kato, M. J. Zhang, M. Wang, L. Lanting, H. Yuan, J. J. Rossi, and R. Natarajan. 2007. ’MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors’, Proc Natl Acad Sci U S A, 104: 3432-7.
- Keller, A.; Gröger, L.; Tschernig, T.; Solomon, J.; Laham, O.; Schaum, N.; Wagner, V.; Kern, F.; Schmartz, G.P.; Li, Y.; et al. miRNATissueAtlas2: an update to the human miRNA tissue atlas. Nucleic Acids Res. 2021, 50, D211–D221. [Google Scholar] [CrossRef]
- Koga, K.; Yokoi, H.; Mori, K.; Kasahara, M.; Kuwabara, T.; Imamaki, H.; Ishii, A.; Mori, K.P.; Kato, Y.; Ohno, S.; et al. MicroRNA-26a inhibits TGF-β-induced extracellular matrix protein expression in podocytes by targeting CTGF and is downregulated in diabetic nephropathy. Diabetologia 2015, 58, 2169–2180. [Google Scholar] [CrossRef]
- Kolde, Raivo. 2019. ’pheatmap: Pretty Heatmaps. ’, R package version 1.0.12. Available online: https://CRAN.R-project.org/package=pheatmap.
- Kozera, B. , and M. Rapacz. 2013. ’Reference genes in real-time PCR’, J Appl Genet, 54: 391-406.
- Krupa, A.; Jenkins, R.; Luo, D.D.; Lewis, A.; Phillips, A.; Fraser, D. Loss of MicroRNA-192 Promotes Fibrogenesis in Diabetic Nephropathy. J. Am. Soc. Nephrol. 2010, 21, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Kölling, M.; Kaucsar, T.; Schauerte, C.; Hübner, A.; Dettling, A.; Park, J.-K.; Busch, M.; Wulff, X.; Meier, M.; Scherf, K.; et al. Therapeutic miR-21 Silencing Ameliorates Diabetic Kidney Disease in Mice. Mol. Ther. 2016, 25, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Lai, J. Y. J. Luo, C. O’Connor, X. Jing, V. Nair, W. Ju, A. Randolph, I. Z. Ben-Dov, R. N. Matar, D. Briskin, J. Zavadil, R. G. Nelson, T. Tuschl, F. C. Brosius, 3rd, M. Kretzler, and M. Bitzer. 2015. ’MicroRNA-21 in glomerular injury’, J Am Soc Nephrol, 26: 805-16.
- Lee, H.W.; Khan, S.Q.; Khaliqdina, S.; Altintas, M.M.; Grahammer, F.; Zhao, J.L.; Koh, K.H.; Tardi, N.J.; Faridi, M.H.; Geraghty, T.; et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-regulation of ErbB4 and Notch-1. J. Biol. Chem. 2017, 292, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lu, Z.; Jia, J.; Zheng, Z.; Lin, S. MiR-124 is Related to Podocytic Adhesive Capacity Damage in STZ-Induced Uninephrectomized Diabetic Rats. Kidney Blood Press. Res. 2013, 37, 422–431. [Google Scholar] [CrossRef]
- Li, H.; Zhu, X.; Zhang, J.; Shi, J. MicroRNA-25 inhibits high glucose-induced apoptosis in renal tubular epithelial cells via PTEN/AKT pathway. Biomed. Pharmacother. 2017, 96, 471–479. [Google Scholar] [CrossRef]
- Li, N. L. J. Wang, W. L. Xu, S. Liu, and J. Y. Yu. 2019. ’MicroRNA-379-5p suppresses renal fibrosis by regulating the LIN28/let-7 axis in diabetic nephropathy’, Int J Mol Med, 44: 1619-28.
- Li, X.; Dong, Z.-Q.; Chang, H.; Zhou, H.-B.; Wang, J.; Yang, Z.-J.; Qiu, M.; Bai, W.-F.; Shi, S.-L. Screening and identification of key microRNAs and regulatory pathways associated with the renal fibrosis process. Mol. Omics 2022, 18, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Y.-H.; Li, F.; Yang, T.; Lu, Y.W.; Geng, Y.-J. microRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009, 381, 81–83. [Google Scholar] [CrossRef]
- Liang, Q. J. He, Q. Yang, Q. Zhang, and Y. Xu. 2022. ’MicroRNA-335-5p alleviates inflammatory response, airway fibrosis, and autophagy in childhood asthma through targeted regulation of autophagy related 5’, Bioengineered, 13: 1791-801.
- Lin, C.-L.; Lee, P.-H.; Hsu, Y.-C.; Lei, C.-C.; Ko, J.-Y.; Chuang, P.-C.; Huang, Y.-T.; Wang, S.-Y.; Wu, S.-L.; Chen, Y.-S.; et al. MicroRNA-29a Promotion of Nephrin Acetylation Ameliorates Hyperglycemia-Induced Podocyte Dysfunction. J. Am. Soc. Nephrol. 2014, 25, 1698–1709. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Liu, S.; Li, H.; Yuan, X.; Feng, B.; Bai, H.; Zhao, B.; Chu, Y.; Li, H. Effects and mechanism of miR-23b on glucose-mediated epithelial-to-mesenchymal transition in diabetic nephropathy. Int. J. Biochem. Cell Biol. 2016, 70, 149–160. [Google Scholar] [CrossRef]
- Liu, W.-T.; Peng, F.-F.; Li, H.-Y.; Chen, X.-W.; Gong, W.-Q.; Chen, W.-J.; Chen, Y.-H.; Li, P.-L.; Li, S.-T.; Xu, Z.-Z.; et al. Metadherin facilitates podocyte apoptosis in diabetic nephropathy. Cell Death Dis. 2016, 7, e2477–e2477. [Google Scholar] [CrossRef]
- Liu, X.-D.; Zhang, L.-Y.; Zhu, T.-C.; Zhang, R.-F.; Wang, S.-L.; Bao, Y. Overexpression of miR-34c inhibits high glucose-induced apoptosis in podocytes by targeting Notch signaling pathways. Int J Clin Exp Pathol 2015, 8, 4525–34. [Google Scholar] [PubMed]
- Liu, Y.; Gao, G.; Yang, C.; Zhou, K.; Shen, B.; Liang, H.; Jiang, X. Stability of miR-126 in Urine and Its Potential as a Biomarker for Renal Endothelial Injury with Diabetic Nephropathy. Int. J. Endocrinol. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Liu, J.; Han, P.; Li, X.; Bai, H.; Zhang, C.; Sun, X.; Teng, Y.; Zhang, Y.; et al. Variations in MicroRNA-25 Expression Influence the Severity of Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2017, 28, 3627–3638. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, B.; Liu, Y.; Chen, S.; Yang, J.; Liu, J.; Sun, G.; Bei, W.-J.; Wang, K.; Chen, Z.; et al. MicroRNA expression profile by next-generation sequencing in a novel rat model of contrast-induced acute kidney injury. Ann. Transl. Med. 2019, 7, 178–178. [Google Scholar] [CrossRef]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.J.; Danesh, F.R. Identification of MicroRNA-93 as a Novel Regulator of Vascular Endothelial Growth Factor in Hyperglycemic Conditions. J. Biol. Chem. 2010, 285, 23457–23465. [Google Scholar] [CrossRef] [PubMed]
- MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy’, J Biol Chem, 286: 11837-48.
- López-Guerrero, José A., Mar Valés-Gómez, Francesc E. Borrás, Juan Manuel Falcón-Pérez, María J. Vicent, and María Yáñez-Mó. 2023. ’Standardising the preanalytical reporting of biospecimens to improve reproducibility in extracellular vesicle research – A GEIVEX study’, Journal of Extracellular Biology, 2: e76.
- Love, M. I. W. Huber, and S. Anders. 2014. ’Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2’, Genome Biol, 15: 550.
- Margolis, E.; Brown, G.; Partin, A.; Carter, B.; McKiernan, J.; Tutrone, R.; Torkler, P.; Fischer, C.; Tadigotla, V.; Noerholm, M.; et al. Predicting high-grade prostate cancer at initial biopsy: clinical performance of the ExoDx (EPI) Prostate Intelliscore test in three independent prospective studies. Prostate Cancer Prostatic Dis. 2021, 25, 296–301. [Google Scholar] [CrossRef]
- Mateescu, B. E. J. Kowal, B. W. van Balkom, S. Bartel, S. N. Bhattacharyya, E. I. Buzás, A. H. Buck, P. de Candia, F. W. Chow, S. Das, T. A. Driedonks, L. Fernández-Messina, F. Haderk, A. F. Hill, J. C. Jones, K. R. Van Keuren-Jensen, C. P. Lai, C. Lässer, I. D. Liegro, T. R. Lunavat, M. J. Lorenowicz, S. L. Maas, I. Mäger, M. Mittelbrunn, S. Momma, K. Mukherjee, M. Nawaz, D. M. Pegtel, M. W. Pfaffl, R. M. Schiffelers, H. Tahara, C. Théry, J. P. Tosar, M. H. Wauben, K. W. Witwer, and E. N. Nolte-’t Hoen. 2017. ’Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper’, J Extracell Vesicles, 6: 1286095.
- McClelland, A.D.; Herman-Edelstein, M.; Komers, R.; Jha, J.C.; Winbanks, C.E.; Hagiwara, S.; Gregorevic, P.; Kantharidis, P.; Cooper, M.E. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin. Sci. 2015, 129, 1237–1249. [Google Scholar] [CrossRef]
- Miller, D.; Eagle-Hemming, B.; Sheikh, S.; Joel-David, L.; Adebayo, A.; Lai, F.Y.; Roman, M.; Kumar, T.; Aujla, H.; Murphy, G.J.; et al. Urinary extracellular vesicles and micro-RNA as markers of acute kidney injury after cardiac surgery. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Min, K.-H.; Yang, W.-M.; Lee, W. Saturated fatty acids-induced miR-424–5p aggravates insulin resistance via targeting insulin receptor in hepatocytes. Biochem. Biophys. Res. Commun. 2018, 503, 1587–1593. [Google Scholar] [CrossRef]
- Miranda, K.C.; Bond, D.T.; Levin, J.Z.; Adiconis, X.; Sivachenko, A.; Russ, C.; Brown, D.; Nusbaum, C.; Russo, L.M. Massively Parallel Sequencing of Human Urinary Exosome/Microvesicle RNA Reveals a Predominance of Non-Coding RNA. PLOS ONE 2014, 9, e96094. [Google Scholar] [CrossRef]
- Miranda, K. C. D. T. Bond, M. McKee, J. Skog, T. G. Păunescu, N. Da Silva, D. Brown, and L. M. Russo. 2010. ’Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease’, Kidney Int, 78: 191-9.
- Mishra, A.; Ayasolla, K.; Kumar, V.; Lan, X.; Vashistha, H.; Aslam, R.; Hussain, A.; Chowdhary, S.; Shoshtari, S.M.; Paliwal, N.; et al. Modulation of apolipoprotein L1-microRNA-193a axis prevents podocyte dedifferentiation in high-glucose milieu. Am. J. Physiol. Physiol. 2018, 314, F832–F843. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Singh, R.S.; Kumari, M.; Garg, D.; Upadhyay, A.; Ecelbarger, C.M.; Tripathy, S.; Tiwari, S. Urinary Exosomal microRNA-451-5p Is a Potential Early Biomarker of Diabetic Nephropathy in Rats. PLOS ONE 2016, 11, e0154055–e0154055. [Google Scholar] [CrossRef] [PubMed]
- Mussack, V.; Wittmann, G.; Pfaffl, M.W. Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing—Enabling robust and non-invasive biomarker research. Biomol. Detect. Quantif. 2019, 17, 100089. [Google Scholar] [CrossRef] [PubMed]
- Müller-Deile, J.; Dannenberg, J.; Schroder, P.; Lin, M.-H.; Miner, J.H.; Chen, R.; Bräsen, J.-H.; Thum, T.; Nyström, J.; Staggs, L.B.; et al. Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int. 2017, 92, 836–849. [Google Scholar] [CrossRef]
- Nieuwland, R.; Siljander, P.R.-M.; Falcón-Pérez, J.M.; Witwer, K.W. Reproducibility of extracellular vesicle research. Eur. J. Cell Biol. 2022, 101, 151226. [Google Scholar] [CrossRef]
- Oh, H. J. M. Kato, S. Deshpande, E. Zhang, S. Das, L. Lanting, M. Wang, and R. Natarajan. 2016. ’Inhibition of the processing of miR-25 by HIPK2-Phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy’, Sci Rep, 6: 38789.
- Oosthuyzen, W.; Sime, N.E.L.; Ivy, J.R.; Turtle, E.J.; Street, J.M.; Pound, J.; Bath, L.E.; Webb, D.J.; Gregory, C.D.; Bailey, M.; et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J. Physiol. 2013, 591, 5833–5842. [Google Scholar] [CrossRef]
- Park, J.T.; Kato, M.; Yuan, H.; Castro, N.; Lanting, L.; Wang, M.; Natarajan, R. FOG2 Protein Down-regulation by Transforming Growth Factor-β1-induced MicroRNA-200b/c Leads to Akt Kinase Activation and Glomerular Mesangial Hypertrophy Related to Diabetic Nephropathy. J. Biol. Chem. 2013, 288, 22469–22480. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, O.-H.; Lee, K.; Park, I.B.; Kim, N.H.; Moon, S.; Im, J.; Sharma, S.P.; Oh, B.-C.; Nam, S.; et al. Plasma and urinary extracellular vesicle microRNAs and their related pathways in diabetic kidney disease. Genomics 2022, 114, 110407. [Google Scholar] [CrossRef]
- Park, S.; Lee, K.; Park, I.B.; Kim, N.H.; Cho, S.; Rhee, W.J.; Oh, Y.; Choi, J.; Nam, S.; Lee, D.H. The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Res. Clin. Pr. 2020, 160, 108010. [Google Scholar] [CrossRef]
- Pavkovic, M.; Vaidya, V.S. MicroRNAs and drug-induced kidney injury. Pharmacol. Ther. 2016, 163, 48–57. [Google Scholar] [CrossRef]
- Pruneda, J. N. F. D. Smith, A. Daurie, D. L. Swaney, J. Villén, J. D. Scott, A. W. Stadnyk, I. Le Trong, R. E. Stenkamp, R. E. Klevit, J. R. Rohde, and P. S. Brzovic. 2014. ’E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis’, Embo j, 33: 437-49.
- Puhka, M. L. Thierens, D. Nicorici, T. Forsman, T. Mirtti, T. Af Hällström, E. Serkkola, and A. Rannikko. 2022. ’Exploration of Extracellular Vesicle miRNAs, Targeted mRNAs and Pathways in Prostate Cancer: Relation to Disease Status and Progression’, Cancers (Basel), 14.
- Qian, X.; Tan, J.; Liu, L.; Chen, S.; You, N.; Yong, H.; Pan, M.; You, Q.; Ding, D.; Lu, Y. MicroRNA-134-5p promotes high glucose-induced podocyte apoptosis by targeting bcl-2. Am J Transl Res 2018, 10, 989–997. [Google Scholar] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Théry, C.; Falcón-Pérez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
- Sáenz-Cuesta, M.; Arbelaiz, A.; Oregi, A.; Irizar, H.; Osorio-Querejeta, I.; Muñoz-Culla, M.; Banales, J.M.; Falcón-Pérez, J.M.; Olascoaga, J.; Otaegui, D. Methods for extracellular vesicles isolation in a hospital setting. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef]
- Scian, M.J.; Maluf, D.G.; David, K.G.; Archer, K.J.; Suh, J.L.; Wolen, A.R.; Mba, M.U.; Massey, H.D.; King, A.L.; Gehr, T.; et al. MicroRNA Profiles in Allograft Tissues and Paired Urines Associate With Chronic Allograft Dysfunction With IF/TA. Am. J. Transplant. 2011, 11, 2110–2122. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Cipolloni, L.; Messina, G.; Aromatario, M.; Polo, L.; Turillazzi, E.; Pomara, C. miRNAs as Novel Biomarkers of Chronic Kidney Injury in Anabolic-Androgenic Steroid Users: An Experimental Study. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Shin, Y. D. Y. Kim, J. Y. Ko, Y. M. Woo, and J. H. Park. 2018. ’Regulation of KLF12 by microRNA-20b and microRNA-106a in cystogenesis’, Faseb j, 32: 3574-82.
- Singh, A. D. S. Patnam, R. Koyyada, R. Samal, S. B. Alvi, G. Satyanaryana, R. Andrews, A. K. Panigrahi, A. K. Rengan, S. S. Mudigonda, S. Maitra, and M. V. Sasidhar. 2022. ’Identifying stable reference genes in polyethene glycol precipitated urinary extracellular vesicles for RT-qPCR-based gene expression studies in renal graft dysfunction patients’, Transpl Immunol, 75: 101715.
- Song, L.; Feng, S.; Yu, H.; Shi, S. Dexmedetomidine Protects Against Kidney Fibrosis in Diabetic Mice by Targeting miR-101-3p-Mediated EndMT. Dose-Response 2022, 20. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Yeri, A.; Cheah, P.S.; Chung, A.; Danielson, K.; De Hoff, P.; Filant, J.; Laurent, C.D.; Laurent, L.D.; Magee, R.; et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 2019, 177, 446–462. [Google Scholar] [CrossRef]
- Su, J. J. Ren, H. Chen, and B. Liu. 2020. ’MicroRNA-140-5p ameliorates the high glucose-induced apoptosis and inflammation through suppressing TLR4/NF-κB signaling pathway in human renal tubular epithelial cells’, Biosci Rep, 40.
- Sun, J.; Li, Z.P.; Zhang, R.Q.; Zhang, H.M. Repression of miR-217 protects against high glucose-induced podocyte injury and insulin resistance by restoring PTEN-mediated autophagy pathway. Biochem. Biophys. Res. Commun. 2017, 483, 318–324. [Google Scholar] [CrossRef]
- Sun, Y. R. Peng, H. Peng, H. Liu, L. Wen, T. Wu, H. Yi, A. Li, and Z. Zhang. 2016. ’miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy’, Mol Cell Endocrinol, 433: 75-86.
- Sun, Z.; Ma, Y.; Chen, F.; Wang, S.; Chen, B.; Shi, J. miR-133b and miR-199b knockdown attenuate TGF-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting SIRT1 in diabetic nephropathy. Eur. J. Pharmacol. 2018, 837, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D. A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen, and C. V. Mering. 2019. ’STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets’, Nucleic Acids Res, 47: D607-d13.
- Toth-Manikowski, S.; Atta, M.G. Diabetic Kidney Disease: Pathophysiology and Therapeutic Targets. J. Diabetes Res. 2015, 2015, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Trevisani, F.; Ghidini, M.; Larcher, A.; Lampis, A.; Lote, H.; Manunta, P.; Alibrandi, M.T.S.; Zagato, L.; Citterio, L.; Dell’Antonio, G.; et al. MicroRNA 193b-3p as a predictive biomarker of chronic kidney disease in patients undergoing radical nephrectomy for renal cell carcinoma. Br. J. Cancer 2016, 115, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-C.; Kuo, M.-C.; Hung, W.-W.; Wu, L.-Y.; Wu, P.-H.; Chang, W.-A.; Kuo, P.-L.; Hsu, Y.-L. High Glucose Induces Mesangial Cell Apoptosis through miR-15b-5p and Promotes Diabetic Nephropathy by Extracellular Vesicle Delivery. Mol. Ther. 2020, 28, 963–974. [Google Scholar] [CrossRef]
- Tuttle, K.R.; Agarwal, R.; Alpers, C.E.; Bakris, G.L.; Brosius, F.C.; Kolkhof, P.; Uribarri, J. Molecular mechanisms and therapeutic targets for diabetic kidney disease. Kidney Int. 2022, 102, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al.; The UniProt Consortium UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef]
- Vago, R.; Radano, G.; Zocco, D.; Zarovni, N. Urine stabilization and normalization strategies favor unbiased analysis of urinary EV content. Sci. Rep. 2022, 12, 1–20. [Google Scholar] [CrossRef]
- EV-TRACK Consortium; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- van Royen, M. E. C. Soekmadji, C. Grange, J. P. Webber, T. Tertel, M. Droste, A. Buescher, B. Giebel, G. W. Jenster, A. Llorente, C. J. Blijdorp, D. Burger, U. Erdbrügger, and E. S. Martens-Uzunova. 2023. ’The quick reference card "Storage of urinary EVs" - A practical guideline tool for research and clinical laboratories’, J Extracell Vesicles, 12: e12286.
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Wan, R.J.; Li, Y.H. MicroRNA-146a/NAPDH oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol. Med. Rep. 2018, 17, 4759–4766. [Google Scholar] [CrossRef] [PubMed]
- Wang, B. M. Herman-Edelstein, P. Koh, W. Burns, K. Jandeleit-Dahm, A. Watson, M. Saleem, G. J. Goodall, S. M. Twigg, M. E. Cooper, and P. Kantharidis. 2010. ’E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta’, Diabetes, 59: 1794-802.
- Wang, B.; Jha, J.C.; Hagiwara, S.; McClelland, A.D.; Jandeleit-Dahm, K.; Thomas, M.C.; Cooper, M.E.; Kantharidis, P. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014, 85, 352–361. [Google Scholar] [CrossRef]
- Wang, B.; Koh, P.; Winbanks, C.; Coughlan, M.T.; McClelland, A.; Watson, A.; Jandeleit-Dahm, K.; Burns, W.C.; Thomas, M.C.; Cooper, M.E.; et al. miR-200a Prevents Renal Fibrogenesis Through Repression of TGF-β2 Expression. Diabetes 2010, 60, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of microRNA-29 Expression by TGF-β1 Promotes Collagen Expression and Renal Fibrosis. J. Am. Soc. Nephrol. 2012, 23, 252–265. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, F.; Tian, Q.; Sheng, K. CircVMA21 ameliorates lipopolysaccharide (LPS)-induced HK-2 cell injury depending on the regulation of miR-7-5p/PPARA. Autoimmunity 2021, 55, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.-H.; Lai, F.M.-M.; Chow, K.-M.; Li, P.K.-T.; Szeto, C.-C. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin. Chim. Acta 2013, 418, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Duan, L.; Tian, L.; Liu, J.; Wang, S.; Gao, Y.; Yang, J. Serum miR-21 may be a Potential Diagnostic Biomarker for Diabetic Nephropathy. Exp. Clin. Endocrinol. Diabetes 2015, 124, 417–423. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Gao, Y.-B.; Zhang, N.; Zou, D.-W.; Wang, P.; Zhu, Z.-Y.; Li, J.-Y.; Zhou, S.-N.; Wang, S.-C.; Wang, Y.-Y.; et al. miR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol. Cell. Endocrinol. 2014, 392, 163–172. [Google Scholar] [CrossRef]
- Wang, L.; Li, H. MiR-770-5p facilitates podocyte apoptosis and inflammation in diabetic nephropathy by targeting TIMP3. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef]
- Wang, S. X. Wen, X. R. Han, Y. J. Wang, M. Shen, S. H. Fan, J. Zhuang, Z. F. Zhang, Q. Shan, M. Q. Li, B. Hu, C. H. Sun, D. M. Wu, J. Lu, and Y. L. Zheng. 2018. ’Repression of microRNA-382 inhibits glomerular mesangial cell proliferation and extracellular matrix accumulation via FoxO1 in mice with diabetic nephropathy’, Cell Prolif, 51: e12462.
- Wang, X.; Lin, B.; Nie, L.; Li, P. microRNA-20b contributes to high glucose-induced podocyte apoptosis by targeting SIRT7. Mol. Med. Rep. 2017, 16, 5667–5674. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, E.; Wang, Y.; Jiang, Z.; Gui, D.; Cheng, D.; Chen, T.; Wang, N. MiR-196a Regulates High Glucose-Induced Mesangial Cell Hypertrophy by Targeting p27kip1. JALA: J. Assoc. Lab. Autom. 2015, 20, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, E.; Wang, Y.; Li, J.; Cheng, D.; Chen, Y.; Gui, D.; Wang, N. Cross talk between miR-214 and PTEN attenuates glomerular hypertrophy under diabetic conditions. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Zhang, L.; Bai, L.; Chen, S.; Wu, H.; Sun, L.; Wang, X. miR-30b-5p modulate renal epithelial-mesenchymal transition in diabetic nephropathy by directly targeting SNAI1. Biochem. Biophys. Res. Commun. 2021, 535, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Z. J. Zheng, Y. J. Jia, Y. L. Yang, and Y. M. Xue. 2018. ’Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease’, J Transl Med, 16: 146.
- Wei, B. Y. S. Liu, and H. X. Guan. 2020. ’MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes’, Exp Ther Med, 19: 1915-24.
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York).
- Wickham, H. Reshaping Data with thereshapePackage. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Wu, L. Q. Wang, F. Guo, X. Ma, H. Ji, F. Liu, Y. Zhao, and G. Qin. 2016. ’MicroRNA-27a Induces Mesangial Cell Injury by Targeting of PPARγ, and its In Vivo Knockdown Prevents Progression of Diabetic Nephropathy’, Sci Rep, 6: 26072.
- Xie, Y.; Jia, Y.; Cuihua, X.; Hu, F.; Xue, M.; Xue, Y. Urinary Exosomal MicroRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J. Diabetes Res. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Xu, H.; Sun, F.; Li, X.; Sun, L. Down-regulation of miR-23a inhibits high glucose-induced EMT and renal fibrogenesis by up-regulation of SnoN. Hum. Cell 2018, 31, 22–32. [Google Scholar] [CrossRef]
- Xu, K.; Liu, Q.; Wu, K.; Liu, L.; Zhao, M.; Yang, H.; Wang, X.; Wang, W. Extracellular vesicles as potential biomarkers and therapeutic approaches in autoimmune diseases. J. Transl. Med. 2020, 18, 1–8. [Google Scholar] [CrossRef]
- Xu, P.; Guan, M.-P.; Bi, J.-G.; Wang, D.; Zheng, Z.-J.; Xue, Y.-M. High glucose down-regulates microRNA-181a-5p to increase pro-fibrotic gene expression by targeting early growth response factor 1 in HK-2 cells. Cell. Signal. 2017, 31, 96–104. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Fan, L.; He, X. miR-423-5p suppresses high-glucose-induced podocyte injury by targeting Nox4. Biochem. Biophys. Res. Commun. 2018, 505, 339–345. [Google Scholar] [CrossRef]
- Xue, M.; Li, Y.; Hu, F.; Jia, Y.-J.; Zheng, Z.-J.; Wang, L.; Xue, Y.-M. High glucose up-regulates microRNA-34a-5p to aggravate fibrosis by targeting SIRT1 in HK-2 cells. Biochem. Biophys. Res. Commun. 2018, 498, 38–44. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Q.; Li, S. MicroRNA-218 promotes high glucose-induced apoptosis in podocytes by targeting heme oxygenase-1. Biochem. Biophys. Res. Commun. 2016, 471, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, Z.; Dong, J.; Sheng, S.; Wang, Y.; Yu, L.; Wang, H.; Tang, L. miR-374a Regulates Inflammatory Response in Diabetic Nephropathy by Targeting MCP-1 Expression. Front. Pharmacol. 2018, 9, 900. [Google Scholar] [CrossRef]
- Yang, Z. Y. Y. Wang, Q. Liu, and M. Wu. 2020. ’microRNA cluster MC-let-7a-1~let-7d promotes autophagy and apoptosis of glioma cells by down-regulating STAT3’, CNS Neurosci Ther, 26: 319-31.
- Yao, T.; Zha, D.; Gao, P.; Shui, H.; Wu, X. MiR-874 alleviates renal injury and inflammatory response in diabetic nephropathy through targeting toll-like receptor-4. J. Cell. Physiol. 2018, 234, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Yekula, A.; Muralidharan, K.; Kang, K.M.; Wang, L.; Balaj, L.; Carter, B.S. From laboratory to clinic: Translation of extracellular vesicle based cancer biomarkers. Methods 2020, 177, 58–66. [Google Scholar] [CrossRef]
- Yu, L.; Gu, T.; Shi, E.; Wang, Y.; Fang, Q.; Wang, C. Dysregulation of renal microRNA expression after deep hypothermic circulatory arrest in rats. Eur. J. Cardio-Thoracic Surg. 2016, 49, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jia, Y.-Y.; Wang, M.; Mu, L.; Li, H.-J. PTGER3 and MMP-2 play potential roles in diabetic nephropathy via competing endogenous RNA mechanisms. BMC Nephrol. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Zanchi, C.; Macconi, D.; Trionfini, P.; Tomasoni, S.; Rottoli, D.; Locatelli, M.; Rudnicki, M.; Vandesompele, J.; Mestdagh, P.; Remuzzi, G.; et al. MicroRNA-184 is a downstream effector of albuminuria driving renal fibrosis in rats with diabetic nephropathy. Diabetologia 2017, 60, 1114–1125. [Google Scholar] [CrossRef]
- Zang, J.; Maxwell, A.P.; Simpson, D.A.; McKay, G.J. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Zha, F.; Bai, L.; Tang, B.; Li, J.; Wang, Y.; Zheng, P.; Ji, T.; Bai, S. MicroRNA-503 contributes to podocyte injury via targeting E2F3 in diabetic nephropathy. J. Cell. Biochem. 2019, 120, 12574–12581. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Y.; Cai, R.; Jin, J.; He, Q. Differential Expression of Urinary Exosomal Small RNAs in Idiopathic Membranous Nephropathy. BioMed Res. Int. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Zhang, L.; He, S.; Guo, S.; Xie, W.; Xin, R.; Yu, H.; Yang, F.; Qiu, J.; Zhang, D.; Zhou, S.; et al. Down-regulation of miR-34a alleviates mesangial proliferation in vitro and glomerular hypertrophy in early diabetic nephropathy mice by targeting GAS1. J. Diabetes Complicat. 2014, 28, 259–264. [Google Scholar] [CrossRef]
- Zhang, R. L. Qin, and J. Shi. 2020. ’MicroRNA-199a-3p suppresses high glucose-induced apoptosis and inflammation by regulating the IKKβ/NF-κB signaling pathway in renal tubular epithelial cells’, Int J Mol Med, 46: 2161-71.
- Zhang, W.; Zhou, X.; Zhang, H.; Yao, Q.; Liu, Y.; Dong, Z. Extracellular vesicles in diagnosis and therapy of kidney diseases. Am. J. Physiol. Physiol. 2016, 311, F844–F851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, S.; Wu, D.; Liu, X.; Shi, M.; Wang, Y.; Zhang, F.; Ding, J.; Xiao, Y.; Guo, B. MicroRNA-22 Promotes Renal Tubulointerstitial Fibrosis by Targeting PTEN and Suppressing Autophagy in Diabetic Nephropathy. J. Diabetes Res. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Peng, H.; Chen, J.; Chen, X.; Han, F.; Xu, X.; He, X.; Yan, N. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009, 583, 2009–2014. [Google Scholar] [CrossRef]
- Zhao, B.; Li, H.; Liu, J.; Han, P.; Zhang, C.; Bai, H.; Yuan, X.; Wang, X.; Li, L.; Ma, H.; et al. MicroRNA-23b Targets Ras GTPase-Activating Protein SH3 Domain-Binding Protein 2 to Alleviate Fibrosis and Albuminuria in Diabetic Nephropathy. J. Am. Soc. Nephrol. 2016, 27, 2597–2608. [Google Scholar] [CrossRef]
- Zhao, D.; Jia, J.; Shao, H. miR-30e targets GLIPR-2 to modulate diabetic nephropathy: in vitro and in vivo experiments. J. Mol. Endocrinol. 2017, 59, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, A.; Guo, F.; Song, Y.; Jing, N.; Ding, X.; Pan, M.; Zhang, H.; Wang, J.; Wu, L.; et al. Urinary Exosomal MiRNA-4534 as a Novel Diagnostic Biomarker for Diabetic Kidney Disease. Front. Endocrinol. 2020, 11. [Google Scholar] [CrossRef]
- Zhong, X.; Chung, A.C.K.; Chen, H.Y.; Dong, Y.; Meng, X.M.; Li, R.; Yang, W.; Hou, F.F.; Lan, H.Y. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013, 56, 663–674. [Google Scholar] [CrossRef]
- Zhou, H. P. S. Yuen, T. Pisitkun, P. A. Gonzales, H. Yasuda, J. W. Dear, P. Gross, M. A. Knepper, and R. A. Star. 2006. ’Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery’, Kidney Int, 69: 1471-6.
- Zhou, X. G. Liu, M. Xu, X. Ying, B. Li, F. Cao, S. Cheng, B. Xiao, M. Cheng, L. Liang, M. Jia, W. Li, J. Liu, and Z. Li. 2022. ’Comprehensive analysis of PTEN-related ceRNA network revealing the key pathways WDFY3-AS2 - miR-21-5p/miR-221-3p/miR-222-3p - TIMP3 as potential biomarker in tumorigenesis and prognosis of kidney renal clear cell carcinoma’, Mol Carcinog, 61: 508-23.
- Zhou, X.; Qu, Z.; Zhu, C.; Lin, Z.; Huo, Y.; Wang, X.; Wang, J.; Li, B. Identification of urinary microRNA biomarkers for detection of gentamicin-induced acute kidney injury in rats. Regul. Toxicol. Pharmacol. 2016, 78, 78–84. [Google Scholar] [CrossRef]
- Zhou, Z.; Wan, J.; Hou, X.; Geng, J.; Li, X.; Bai, X. MicroRNA-27a promotes podocyte injury via PPARγ-mediated β-catenin activation in diabetic nephropathy. Cell Death Dis. 2017, 8, e2658–e2658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Li, H.; Ao, Z.; Xu, H.; Luo, J.; Kaurich, C.; Yang, R.; Zhu, P.-W.; Chen, S.-D.; Wang, X.-D.; et al. Lipidomic identification of urinary extracellular vesicles for non-alcoholic steatohepatitis diagnosis. J. Nanobiotechnology 2022, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zubiri, I.; Posada-Ayala, M.; Sanz-Maroto, A.; Calvo, E.; Martin-Lorenzo, M.; Gonzalez-Calero, L.; de la Cuesta, F.; Lopez, J.A.; Fernandez-Fernandez, B.; Ortiz, A.; et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J. Proteom. 2014, 96, 92–102. [Google Scholar] [CrossRef] [PubMed]
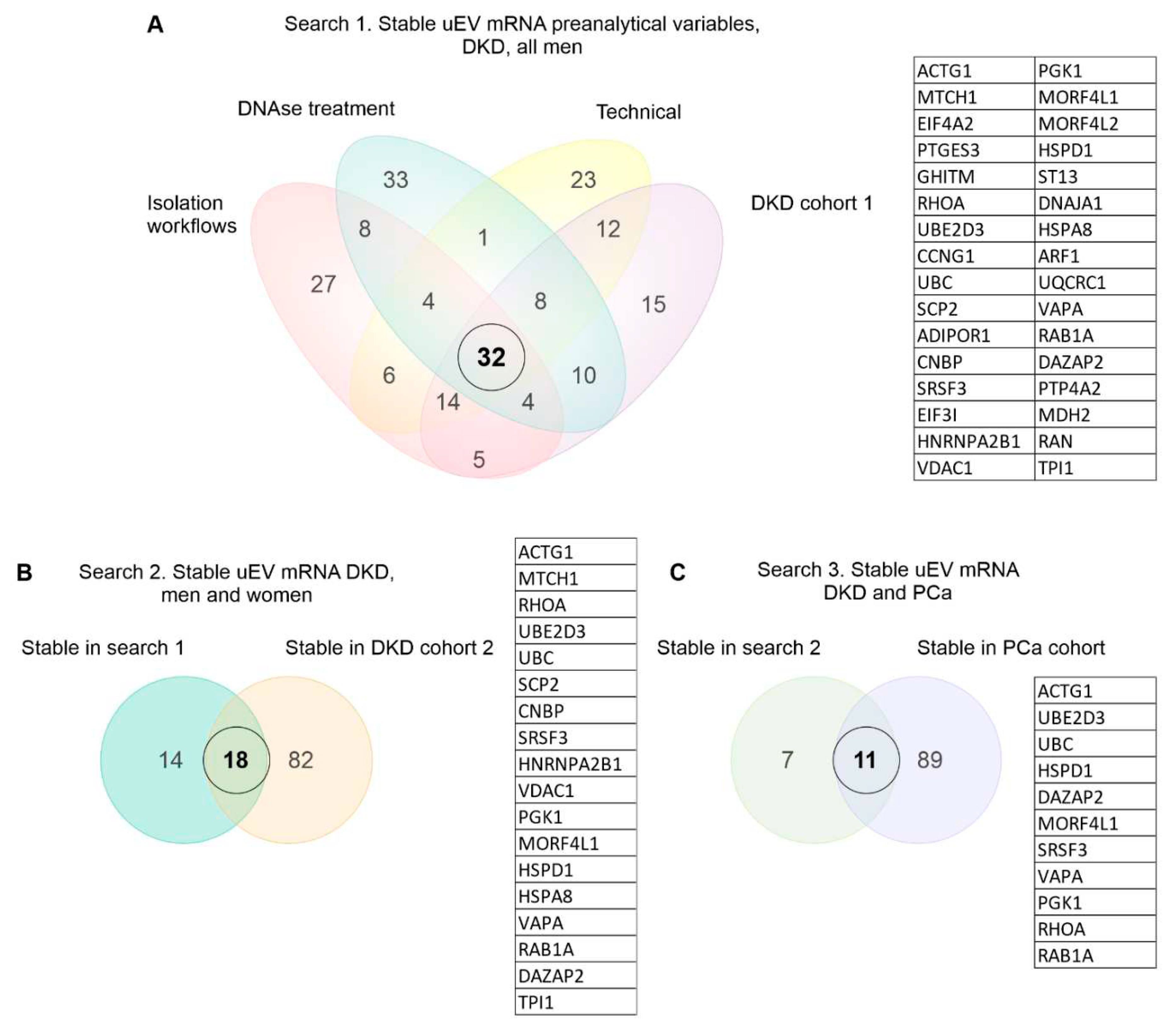
| Study | Description | storage temp | PI* | pre-clearing | isolation method | Urine sample type and disease | n (donors) | Analysis type | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Isolation workflows | uEV were isolated from urines | -80°C | yes | no | UC, HFD, and NG | 24h urine samples from healthy controls and T1D patients with macroalbuminuria. All men (n=20). | healthy controls=5macroalbuminuria=5 | miRNA and mRNA sequencing | (Barreiro et al. 2020) |
| Storage temperature | uEV were isolated from paired urine aliquots stored at -20°C or -80°C. | -80°C/-20°C | yes | no | UC | 24h urine samples from T1D patients with normoalbuminuria, microalbuminuria or macroalbuminuria. All men (n=19). | normoalbuminuria=2macroalbuminuria=2 | miRNA and mRNA sequencing | (Barreiro et al. 2021) |
| DNAse treatment | uEV RNA was isolated adding an in-column DNAse digestion step. | -80°C | yes | no | UC | 24h urine samples from T1D patients with normoalbuminuria, microalbuminuria or macroalbuminuria. All men. | normoalbuminuria=11microalbuminuria=2macroalbuminuria=6 | mRNA sequencing | (Barreiro et al. 2021) |
| ON/24h | uEV were isolated from urines derived from donors that provided on the same day 24h urine (full void during 24 hours) or ON urine (full first void). | -80°C | Yes | no | UC | ON and 24h urine samples from healthy controls and T1D patients with normoalbuminuria or macroalbuminuria. All men. | ON/24 pairs=12 | mRNA sequencing. | (Dwivedi et al. 2023) |
| Pre-clearing | uEV isolated from paired urine aliquots processed +/- pre-clearing before storage. | -80°C | Yes | yes/no | UC | 24h urine samples from T1D patients with microalbuminuria or macroalbuminuria. All men. | pre-clearing pairs=4 | mRNA sequencing | (Dwivedi et al. 2023) |
| Replicability of UC workflow | Pairs of urine aliquots were stored and processed at different time points (up to 5 months) | -80°C | yes | no | UC | 24h urine samples from healthy controls and T1D patients. All men. | duplicates=6triplicates=2 | mRNA sequencing | (Dwivedi et al. 2023) |
| DKD cohort 1 | uEV isolated from urines to | -80°C | yes | yes-no** | UC | 24h or ON urine samples from T1D patients with normoalbuminuria, microalbuminuria or macroalbuminuria. All men. | normoalbuminuria=38microalbuminuria=15macroalbuminuria=19 | mRNA sequencing | (Dwivedi et al. 2023) |
| DKD cohort 2 | uEV isolated from urines to validate candidate markers. | -80°C | yes | yes | UC | 24h urine samples from T1D patients with normoalbuminuria, microalbuminuria or macroalbuminuria. All women. | normoalbuminuria=18microalbuminuria=8macroalbuminuria=4 | mRNA sequencing | unpublished raw count data |
| PCa cohort | uEV isolated from urine samples from PCa patients | -80°C | no | yes | UC | Spot urine samples from healthy technical controls and PCa patients before and after radical prostatectomy. All men | PCa=3healthy controls= 2 (1 men with 3 technical replicates and 1 woman) | mRNA sequencing | (Puhka et al. 2022) |
| raw counts | ||||
|---|---|---|---|---|
| ID | -80°C | -20°C | Association with kidney diseases | |
| Downregulated | hsa-miR-21-5p | 42760 ± 22321 | 230 ± 115 | Downregulation in kidneys from CKD donors. (Sessa et al. 2020) |
| hsa-miR-375 | 11496 ± 4880 | 26 ± 13 | Pro-apoptotic in an in vitro model of AKI (renal tubular cells) (Jones et al. 2018) . | |
| hsa-miR-192-5p | 10651 ± 5178 | 15 ± 8 | Downregulation in DKD. Associated with fibrosis (Krupa et al. 2010). | |
| hsa-miR-378a-3p | 1445 ± 740 | 10 ± 5 | Upregulation seen in biopsies from donors with glomerular diseases (Müller-Deile et al. 2017). | |
| hsa-miR-101-3p | 971 ± 483 | 0 | Downregulated in kidneys from a mouse diabetic nephropathy model (STZ). Antifibrotic (Song et al. 2022). | |
| hsa-miR-107 | 700 ± 270 | 1 ± 0.3 | Downregulated in kidney biopsies from allograft dysfunction (Scian et al. 2011). | |
| hsa-miR-320b | 466 ± 262 | 2 ± 1 | ||
| hsa-miR-345-5p | 236 ± 121 | 0 | Upregulated in urine from a chemical model of AKI in rats (Zhou et al. 2016). | |
| hsa-miR-328-3p | 203 ± 88 | 0 | Downregulated in proximal tubule cells that underwent ischemia/reperfusion (Gao et al. 2022). | |
| hsa-miR-204-3p | 202 ± 157 | 0 | Upregulation protected podocytes exposed to high glucose from apoptosis (Han et al. 2019). | |
| hsa-miR-7-5p | 174 ± 119 | 0 | Downregulation protected proximal tubule cells from LPS in vitro (Wang et al. 2022). | |
| hsa-miR-197-3p | 154 ± 92 | 0 | Downregulated in urine from donors with intermittent MA (Ghai et al. 2018). | |
| hsa-miR-20b-5p | 151 ± 80 | 0 | Downregulated in kidneys and cell lines from mouse models of polycystic kidney disease (Shin et al. 2018). | |
| hsa-miR-148a-5p | 133 ± 60 | 0 | Increased in urine from donors with persistent macroalbuminuria (Ghai et al. 2018). | |
| hsa-miR-10a-3p | 114 ± 79 | 0 | Downregulated in kidneys from a mouse model of AKI (Yu et al. 2016). | |
| hsa-miR-629-5p | 109 ± 81 | 0 | Upregulated in kidney biopsies from donors with acute tubular necrosis (Pavkovic and Vaidya 2016). | |
| hsa-miR-92a-1-5p | 101 ± 59 | 0 | ||
| hsa-miR-193b-3p | 100 ± 62 | 0 | Upregulated in kidney from chronic kidney disease biopsies (Trevisani et al. 2016). | |
| hsa-miR-340-5p | 99 ± 36 | 0 | ||
| hsa-miR-3065-5p | 98 ± 43 | 0 | Upregulated in a mouse model of renal fibrosis (Li et al. 2022). | |
| hsa-miR-106a-5p | 92 ± 50 | 0 | Downregulation associated with podocyte injury induced by high glucose (He and Zeng 2020). | |
| hsa-miR-7704 | 87 ± 37 | 0 | ||
| hsa-miR-324-5p | 74 ± 42 | 0 | ||
| hsa-miR-374b-5p | 60 ± 24 | 0 | Downregulated in diabetic kidney biopsies (Yu et al. 2021). | |
| hsa-miR-99b-3p | 59 ± 19 | 0 | ||
| hsa-miR-4728-3p | 59 ± 20 | 0 | ||
| hsa-miR-132-3p | 57 ± 12 | 0 | Upregulation increases fibrosis in mouse and in vitro (Bijkerk et al. 2016). | |
| hsa-miR-361-5p | 55 ± 15 | 0 | ||
| hsa-miR-664a-5p | 54 ± 15 | 0 | Upregulated in uEV from donors with Idiopathic Membranous Nephropathy (Zhang et al. 2020). | |
| upregulated | hsa-miR-10a-5p | 47380 ± 33187 | 5272 ± 2636 | Downregulated in urine of individuals with AKI (Miller et al. 2022). |
| hsa-miR-125a-5p | 1017 ± 399 | 103 ± 51 | Downregulated in urine from donors with membranous nephropathy (Hou et al. 2021) | |
| hsa-miR-92b-3p | 864 ± 413 | 58 ± 29 | Upregulated in urine from donors with persistent macroalbuminuria (Ghai et al. 2018). | |
| hsa-miR-3960 | 77 ± 44 | 15 ± 7 | Upregulated in kidney biopsies from donors with acute tubular necrosis (Pavkovic and Vaidya 2016) | |
| miRNA (human) | Evidence | miRNA Regulation in disease group | Reference |
|---|---|---|---|
| let-7b-5p | in vitro and in vivo | Down | (Wang, Jha, et al. 2014) |
| miR-15b-5p | in vitro and in vivo | Up | (Tsai et al. 2020) |
| miR-16-5p | in vitro | Down | (Duan et al. 2021) |
| miR-20b-5p | in vitro | Up | (Wang et al. 2017) |
| miR-21-5p | in vitro and in vivo | Down | (Zhang et al. 2009) |
| miR-21-5p | in vitro and in vivo | Up | (Dey et al. 2011) |
| miR-21-5p | in vitro and in vivo | Up | (Zhong et al. 2013) |
| miR-21-5p | in vitro and in vivo | Up | (Chen et al. 2018) |
| miR-21-5p | in vitro and in vivo | - | (Wang, Gao, et al. 2014) |
| miR-21-5p | in vitro and in vivo | Up | (Lai et al. 2015) |
| miR-21-5p | in vitro and in vivo | Up | (Wang, Duan, et al. 2016) |
| miR-21-5p | in vitro and in vivo | Up | (Kölling et al. 2017) |
| miR-21 | in vivo | Up | (McClelland et al. 2015) |
| miR-22 | in vitro and in vivo | Up | (Zhang et al. 2018) |
| miR-23a-3p | in vitro | Up | (Xu, Sun, et al. 2018) |
| miR-23b-3p | in vitro and in vivo | Down | (Liu, Wang, et al. 2016) |
| miR-23b | in vitro and in vivo | Down | (Zhao et al. 2016) |
| miR-25-3p | in vitro and in vivo | Down | (Fu et al. 2010) |
| miR-25-3p | in vitro and in vivo | Down | (Oh et al. 2016) |
| miR-25-3p | in vitro and in vivo | Down | (Li et al. 2017) |
| miR-25 | in vitro and in vivo | Down | (Liu et al. 2017) |
| miR-26a-5p | in vitro and in vivo | Down | (Koga et al. 2015) |
| miR-27a-3p | in vitro and in vivo | Up | (Wu et al. 2016) |
| miR-27a | in vitro and in vivo | Up | (Zhou et al. 2017) |
| miR-29b-3p | in vitro and in vivo | Up | (Beltrami et al. 2018) |
| miR-29b-3p | in vitro and in vivo | Down | (Chen et al. 2014) |
| miR-29c-3p | in vitro and in vivo | Up | (Long et al. 2011) |
| miR-29a | in vitro and in vivo | Down | (Lin et al. 2014) |
| miR-29a | in vitro and in vivo | Down | (Hsu et al. 2016) |
| miR-29a/b/c family | in vitro and in vivo | Down | (Wang et al. 2012) |
| miR-30e-5p | in vitro and in vivo | Down | (Zhao, Jia, and Shao 2017) |
| miR-30s (family) | in vitro and in vivo | Down | (Liu, Peng, et al. 2016) |
| miR-30b-5p | in vitro | Down | (Wang et al. 2021) |
| miR-30c-5p | in vitro and in vivo | Down | (Cui, Yu, and Cui 2020) |
| miR-34a-5p | in vitro and in vivo | Up | (Zhang et al. 2014) |
| miR-34a-5p | in vitro and in vivo | Up | (Xue et al. 2018) |
| miR-34c-5p | in vitro | Down | (Liu et al. 2015) |
| miR-93-5p | in vitro and in vivo | Down | (Long et al. 2010) |
| miR-124-5p | in vivo | Up | (Li et al. 2013) |
| hsa-miR-126-3p | in vitro and in vivo | Up | (Beltrami et al. 2018) |
| miR-130a-3p | in vitro | Down | (Jiang et al. 2018) |
| miR-130b-5p | in vitro and in vivo | Down | (Bai et al. 2016) |
| miR-133b | in vitro and in vivo | Up | (Sun et al. 2018) |
| miR-134-5p | in vitro and in vivo | Up | (Qian et al. 2018) |
| miR-135a-5p | in vitro and in vivo | Up | (He et al. 2014) |
| miR-140-5p | in vitro and in vivo | Down | (Su et al. 2020) |
| miR-145-5p | in vitro and in vivo | Up | (Barutta et al. 2013) |
| miR-145-5p | in vitro | Down | (Wei, Liu, and Guan 2020) |
| miR-146a-5p | in vitro and in vivo | Up | (Huang et al. 2014) |
| miR-146a-5p | in vitro and in vivo | Down | (Lee et al. 2017) |
| miR-146a | in vivo | Down | (Bhatt et al. 2016) |
| miR-146a | in vitro and in vivo | Down | (Wan and Li 2018) |
| miR-155-5p | in vitro and in vivo | Up | (Huang et al. 2014) |
| miR-155-5p | in vitro and in vivo | Up | (Beltrami et al. 2018) |
| miR-155-5p | in vitro | Up | (Wang, Zheng, et al. 2018) |
| miR-181a-5p | in vitro and in vivo | Down | (Xu et al. 2017) |
| miR-192-5p | in vitro and in vivo | Up | (Kato et al. 2007) |
| miR-192-5p | in vitro and in vivo | Down | (Wang et al. 2010) |
| miR-192-5p | in vitro and in vivo | Up | (Kato et al. 2011) |
| miR-192 | in vitro and in vivo | Up in microalbuminuria, Down in macroalbuminuria | (Jia et al. 2016) |
| miR-192 | in vitro and in vivo | Down | (Krupa et al. 2010) |
| miR-192 | in vitro and in vivo | Up | (Deshpande et al. 2013) |
| miR-193a | in vitro | Up | (Mishra et al. 2018) |
| miR-195 | in vitro and in vivo | Down | (Chen et al. 2012) |
| miR-196a-5p | in vitro and in vivo | Down | (Wang et al. 2015) |
| miR-199a-3p | in vitro and in vivo | Down | (Zhang, Qin, and Shi 2020) |
| miR-199b-5p | in vitro and in vivo | Up | (Sun et al. 2018) |
| miR-200a-3p | in vitro and in vivo | Down | (Wang et al. 2011) |
| miR-200 b/c-3p | in vitro and in vivo | Up | (Kato et al. 2011) |
| miR-200 b/c | in vitro and in vivo | Up | (Park et al. 2013) |
| miR-214-3p | in vitro and in vivo | Up | (Wang, Shen, et al. 2016) |
| miR-214-3p | in vitro and in vivo | Up | (Bera et al. 2017) |
| miR-215-5p | in vitro and in vivo | Down | (Wang et al. 2010) |
| miR-216a-5p | in vitro and in vivo | Up | (Kato et al. 2009) |
| miR-216a-5p | in vivo | UP | (Kato et al. 2010) |
| miR-217-5p | in vitro | Up | (Sun et al. 2017) |
| miR-217-5p | in vitro and in vivo | Up | (Kato et al. 2009) |
| miR-218-5p | in vitro | Up | (Yang, Wang, and Li 2016) |
| miR-301a-3p | in vitro | Down | (Jiang et al. 2018) |
| miR-342-3p | in vitro and in vivo | Down | (Jiang et al. 2020) |
| miR-374a | in vitro and in vivo | Down | (Yang et al. 2018) |
| miR-377-3p | in vitro and in vivo | Up | (Wang et al. 2008) |
| miR-379-5p | in vitro and in vivo | Down | (Li et al. 2019) |
| miR-379 megacluster | in vitro and in vivo | Up | (Kato et al. 2016) |
| miR-382 | in vitro and in vivo | Up | (Wang, Wen, et al. 2018) |
| miR-423-5p | in vitro | Down | (Xu, Zhang, et al. 2018) |
| miR-451a | in vitro and in vivo | Down | (Sun et al. 2016) |
| miR-451a | in vivo | Up/Down | (Mohan et al. 2016) |
| miR-503 | in vitro and in vivo | Up | (Zha et al. 2019) |
| miR-770-5p | in vitro and in vivo | Up | (Wang and Li 2020) |
| miR-874 | in vitro and in vivo | Up (overt nephropathy) | (Zanchi et al. 2017) |
| miR-874 | in vitro and in vivo | Down | (Yao et al. 2018) |
| miR-1207-5p | in vitro | Up | (Alvarez et al. 2013) |
| CV | ||||||
|---|---|---|---|---|---|---|
| Isolation workflows | DNAse treatment | Technical datasets | DKD cohort 1 (T1D, men) | DKD cohort 2 (T1D, women) | PCa | |
| HSPD1 | 0.23 | 0.13 | 0.15 | 0.14 | 0.16 | 0.12 |
| SRSF3 | 0.21 | 0.13 | 0.17 | 0.15 | 0.18 | 0.16 |
| VAPA | 0.26 | 0.13 | 0.16 | 0.16 | 0.23 | 0.16 |
| RAB1A | 0.26 | 0.19 | 0.15 | 0.18 | 0.21 | 0.17 |
| MORF4L1 | 0.22 | 0.13 | 0.17 | 0.21 | 0.16 | 0.16 |
| PGK1 | 0.22 | 0.20 | 0.21 | 0.19 | 0.24 | 0.16 |
| RHOA | 0.17 | 0.16 | 0.19 | 0.15 | 0.22 | 0.16 |
| UBE2D3 | 0.18 | 0.14 | 0.13 | 0.15 | 0.20 | 0.11 |
| DAZAP2 | 0.26 | 0.19 | 0.19 | 0.20 | 0.17 | 0.16 |
| UBC | 0.19 | 0.16 | 0.16 | 0.20 | 0.38 | 0.11 |
| ACTG1 | 0.13 | 0.19 | 0.14 | 0.15 | 0.25 | 0.08 |
| GAPDH* | 0.18 | 0.20 | 0.17 | 0.19 | 0.24 | 0.29 |
| UPK1A** | 0.70 | 0.36 | 0.59 | 0.63 | 0.49 | 0.49 |
| Gene name | Entry | Protein names | Gene Ontology (biological process) |
| PGK1 | P00558 | Phosphoglycerate kinase 1 | canonical glycolysis [GO:0061621]; cellular response to hypoxia [GO:0071456]; epithelial cell differentiation [GO:0030855]; gluconeogenesis [GO:0006094]; glycolytic process [GO:0006096]; negative regulation of angiogenesis [GO:0016525]; phosphorylation [GO:0016310]; plasminogen activation [GO:0031639] |
| UBC | P0CG48 | Polyubiquitin-C | modification-dependent protein catabolic process [GO:0019941]; protein ubiquitination [GO:0016567] |
| HSPD1 | P10809 | 60 kDa heat shock protein, mitochondrial | ’de novo’ protein folding [GO:0006458]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic mitochondrial changes [GO:0008637]; B cell activation [GO:0042113]; B cell proliferation [GO:0042100]; biological process involved in interaction with symbiont [GO:0051702]; cellular response to interleukin-7 [GO:0098761]; chaperone-mediated protein complex assembly [GO:0051131]; isotype switching to IgG isotypes [GO:0048291]; mitochondrial unfolded protein response [GO:0034514]; MyD88-dependent toll-like receptor signaling pathway [GO:0002755]; negative regulation of apoptotic process [GO:0043066]; positive regulation of apoptotic process [GO:0043065]; positive regulation of interferon-alpha production [GO:0032727]; positive regulation of interleukin-10 production [GO:0032733]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of macrophage activation [GO:0043032]; positive regulation of T cell activation [GO:0050870]; positive regulation of T cell mediated immune response to tumor cell [GO:0002842]; positive regulation of type II interferon production [GO:0032729]; protein folding [GO:0006457]; protein import into mitochondrial intermembrane space [GO:0045041]; protein maturation [GO:0051604]; protein refolding [GO:0042026]; protein stabilization [GO:0050821]; response to cold [GO:0009409]; response to unfolded protein [GO:0006986]; T cell activation [GO:0042110] |
| UBE2D3 | P61077 | Ubiquitin-conjugating enzyme E2 D3 | apoptotic process [GO:0006915]; DNA repair [GO:0006281]; negative regulation of BMP signaling pathway [GO:0030514]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of protein targeting to mitochondrion [GO:1903955]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein autoubiquitination [GO:0051865]; protein K11-linked ubiquitination [GO:0070979]; protein K48-linked ubiquitination [GO:0070936]; protein modification process [GO:0036211]; protein monoubiquitination [GO:0006513]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567] |
| RHOA | P61586 | Transforming protein RhoA | actin cytoskeleton organization [GO:0030036]; actin cytoskeleton reorganization [GO:0031532]; actin filament organization [GO:0007015]; alpha-beta T cell lineage commitment [GO:0002363]; androgen receptor signaling pathway [GO:0030521]; angiotensin-mediated vasoconstriction involved in regulation of systemic arterial blood pressure [GO:0001998]; aortic valve formation [GO:0003189]; apical junction assembly [GO:0043297]; apolipoprotein A-I-mediated signaling pathway [GO:0038027]; beta selection [GO:0043366]; cell junction assembly [GO:0034329]; cell migration [GO:0016477]; cell-matrix adhesion [GO:0007160]; cellular response to chemokine [GO:1990869]; cellular response to cytokine stimulus [GO:0071345]; cellular response to lipopolysaccharide [GO:0071222]; cerebral cortex cell migration [GO:0021795]; cleavage furrow formation [GO:0036089]; cortical cytoskeleton organization [GO:0030865]; cytoplasmic microtubule organization [GO:0031122]; endothelial cell migration [GO:0043542]; endothelial tube lumen extension [GO:0097498]; establishment of epithelial cell apical/basal polarity [GO:0045198]; establishment or maintenance of cell polarity [GO:0007163]; forebrain radial glial cell differentiation [GO:0021861]; GTP metabolic process [GO:0046039]; kidney development [GO:0001822]; mitotic cleavage furrow formation [GO:1903673]; mitotic spindle assembly [GO:0090307]; motor neuron apoptotic process [GO:0097049]; negative chemotaxis [GO:0050919]; negative regulation of cell migration involved in sprouting angiogenesis [GO:0090051]; negative regulation of cell size [GO:0045792]; negative regulation of cell-substrate adhesion [GO:0010812]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of intracellular steroid hormone receptor signaling pathway [GO:0033144]; negative regulation of motor neuron apoptotic process [GO:2000672]; negative regulation of neuron differentiation [GO:0045665]; negative regulation of neuron projection development [GO:0010977]; negative regulation of oxidative phosphorylation [GO:0090324]; negative regulation of reactive oxygen species biosynthetic process [GO:1903427]; negative regulation of vascular associated smooth muscle cell migration [GO:1904753]; negative regulation of vascular associated smooth muscle cell proliferation [GO:1904706]; neuron migration [GO:0001764]; neuron projection morphogenesis [GO:0048812]; odontogenesis [GO:0042476]; ossification involved in bone maturation [GO:0043931]; positive regulation of actin filament polymerization [GO:0030838]; positive regulation of alpha-beta T cell differentiation [GO:0046638]; positive regulation of cell growth [GO:0030307]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; positive regulation of cytokinesis [GO:0032467]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of leukocyte adhesion to vascular endothelial cell [GO:1904996]; positive regulation of lipase activity [GO:0060193]; positive regulation of neuron apoptotic process [GO:0043525]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of podosome assembly [GO:0071803]; positive regulation of protein serine/threonine kinase activity [GO:0071902]; positive regulation of stress fiber assembly [GO:0051496]; positive regulation of T cell migration [GO:2000406]; positive regulation of translation [GO:0045727]; positive regulation of vascular associated smooth muscle contraction [GO:1904695]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of calcium ion transport [GO:0051924]; regulation of cell migration [GO:0030334]; regulation of cell shape [GO:0008360]; regulation of dendrite development [GO:0050773]; regulation of focal adhesion assembly [GO:0051893]; regulation of microtubule cytoskeleton organization [GO:0070507]; regulation of modification of postsynaptic actin cytoskeleton [GO:1905274]; regulation of modification of postsynaptic structure [GO:0099159]; regulation of neural precursor cell proliferation [GO:2000177]; regulation of osteoblast proliferation [GO:0033688]; regulation of systemic arterial blood pressure by endothelin [GO:0003100]; regulation of transcription by RNA polymerase II [GO:0006357]; response to amino acid [GO:0043200]; response to ethanol [GO:0045471]; response to glucocorticoid [GO:0051384]; response to glucose [GO:0009749]; response to hypoxia [GO:0001666]; response to mechanical stimulus [GO:0009612]; response to xenobiotic stimulus [GO:0009410]; Rho protein signal transduction [GO:0007266]; Roundabout signaling pathway [GO:0035385]; skeletal muscle satellite cell migration [GO:1902766]; skeletal muscle tissue development [GO:0007519]; stress fiber assembly [GO:0043149]; stress-activated protein kinase signaling cascade [GO:0031098]; substantia nigra development [GO:0021762]; substrate adhesion-dependent cell spreading [GO:0034446]; trabecula morphogenesis [GO:0061383]; Wnt signaling pathway, planar cell polarity pathway [GO:0060071]; wound healing, spreading of cells [GO:0044319] |
| RAB1A | P62820 | Ras-related protein Rab-1A | autophagosome assembly [GO:0000045]; autophagy [GO:0006914]; cell migration [GO:0016477]; COPII-coated vesicle cargo loading [GO:0090110]; defense response to bacterium [GO:0042742]; endocytosis [GO:0006897]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; Golgi organization [GO:0007030]; growth hormone secretion [GO:0030252]; intracellular protein transport [GO:0006886]; melanosome transport [GO:0032402]; positive regulation of glycoprotein metabolic process [GO:1903020]; positive regulation of interleukin-8 production [GO:0032757]; substrate adhesion-dependent cell spreading [GO:0034446]; vesicle transport along microtubule [GO:0047496]; vesicle-mediated transport [GO:0016192]; virion assembly [GO:0019068] |
| ACTG1 | P63261 | Actin, cytoplasmic 2 | angiogenesis [GO:0001525]; cellular response to type II interferon [GO:0071346]; maintenance of blood-brain barrier [GO:0035633]; morphogenesis of a polarized epithelium [GO:0001738]; platelet aggregation [GO:0070527]; positive regulation of cell migration [GO:0030335]; positive regulation of gene expression [GO:0010628]; positive regulation of wound healing [GO:0090303]; protein localization to bicellular tight junction [GO:1902396]; regulation of focal adhesion assembly [GO:0051893]; regulation of stress fiber assembly [GO:0051492]; regulation of synaptic vesicle endocytosis [GO:1900242]; regulation of transepithelial transport [GO:0150111]; response to calcium ion [GO:0051592]; response to mechanical stimulus [GO:0009612]; retina homeostasis [GO:0001895]; sarcomere organization [GO:0045214]; tight junction assembly [GO:0120192] |
| SRSF3 | P84103 | Serine/arginine-rich splicing factor 3 (Pre-mRNA-splicing factor SRP20) | cellular response to leukemia inhibitory factor [GO:1990830]; mRNA export from nucleus [GO:0006406]; mRNA splicing, via spliceosome [GO:0000398]; primary miRNA processing [GO:0031053]; regulation of mRNA splicing, via spliceosome [GO:0048024] |
| DAZAP2 | Q15038 | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) | positive regulation of protein serine/threonine kinase activity [GO:0071902]; positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding [GO:1905636]; protein destabilization [GO:0031648]; stress granule assembly [GO:0034063] |
| VAPA | Q9P0L0 | Vesicle-associated membrane protein-associated protein A (VAMP-A) | cell death [GO:0008219]; ceramide transport [GO:0035627]; COPII-coated vesicle budding [GO:0090114]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; membrane fusion [GO:0061025]; negative regulation by host of viral genome replication [GO:0044828]; neuron projection development [GO:0031175]; phospholipid transport [GO:0015914]; positive regulation by host of viral genome replication [GO:0044829]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; protein localization to endoplasmic reticulum [GO:0070972]; sphingomyelin biosynthetic process [GO:0006686]; sterol transport [GO:0015918]; viral release from host cell [GO:0019076] |
| MORF4L1 | Q9UBU8 | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) | chromatin organization [GO:0006325]; double-strand break repair via homologous recombination [GO:0000724]; fibroblast proliferation [GO:0048144]; histone acetylation [GO:0016573]; histone deacetylation [GO:0016575]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; positive regulation of DNA-templated transcription [GO:0045893]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; regulation of apoptotic process [GO:0042981]; regulation of cell cycle [GO:0051726]; regulation of double-strand break repair [GO:2000779] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





