Submitted:
25 May 2023
Posted:
29 May 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Results
Evaluation of the sample
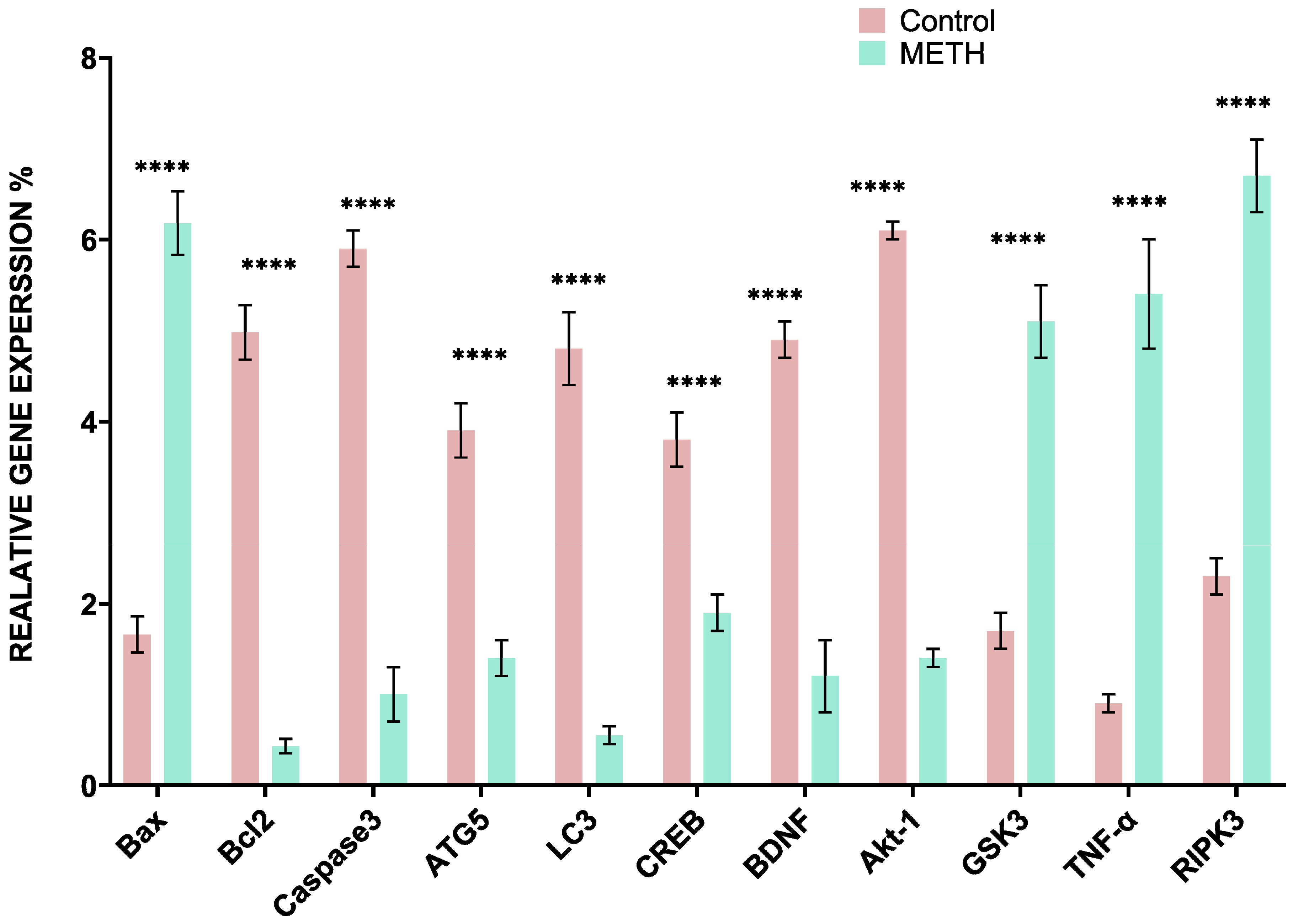
Real-time PCR
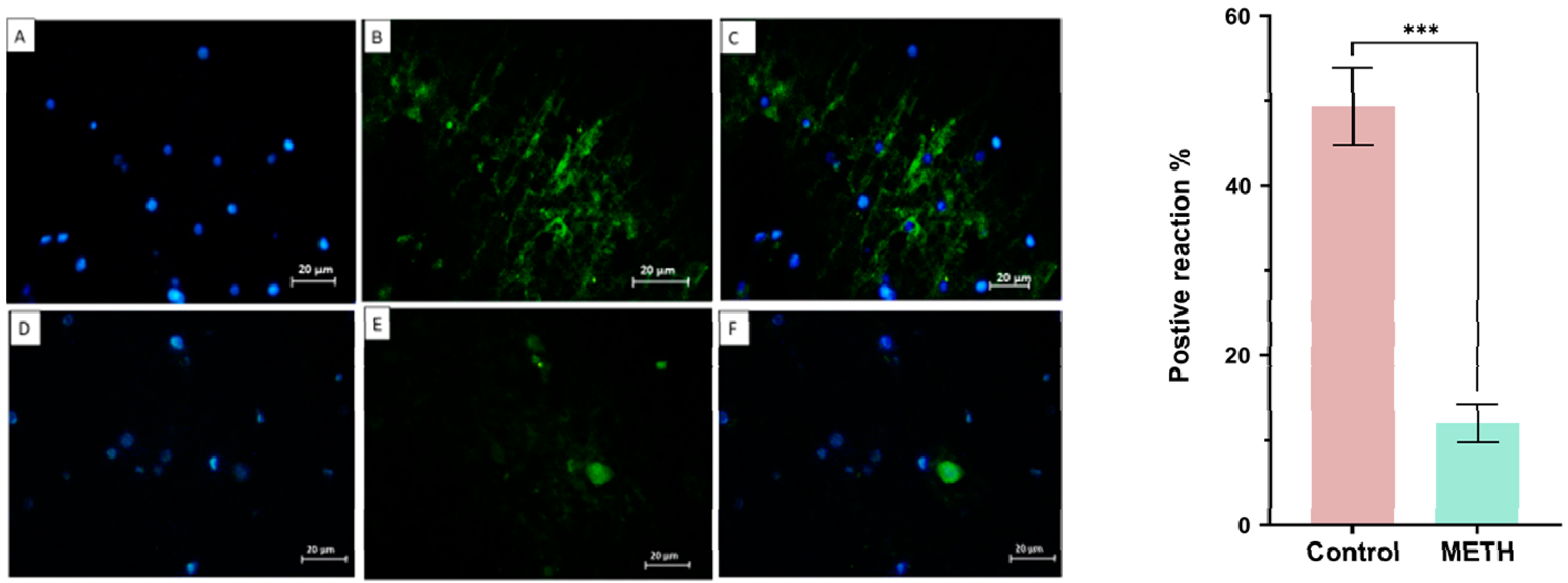
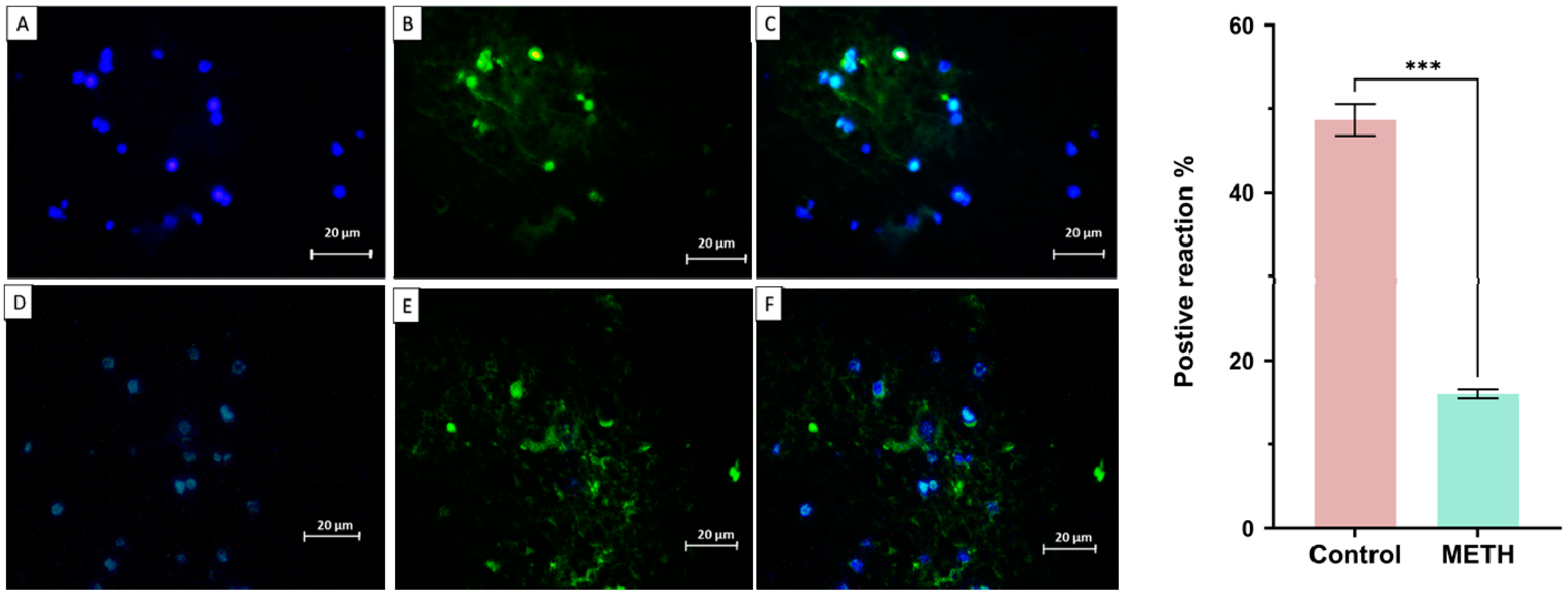
Immunohistochemistry result
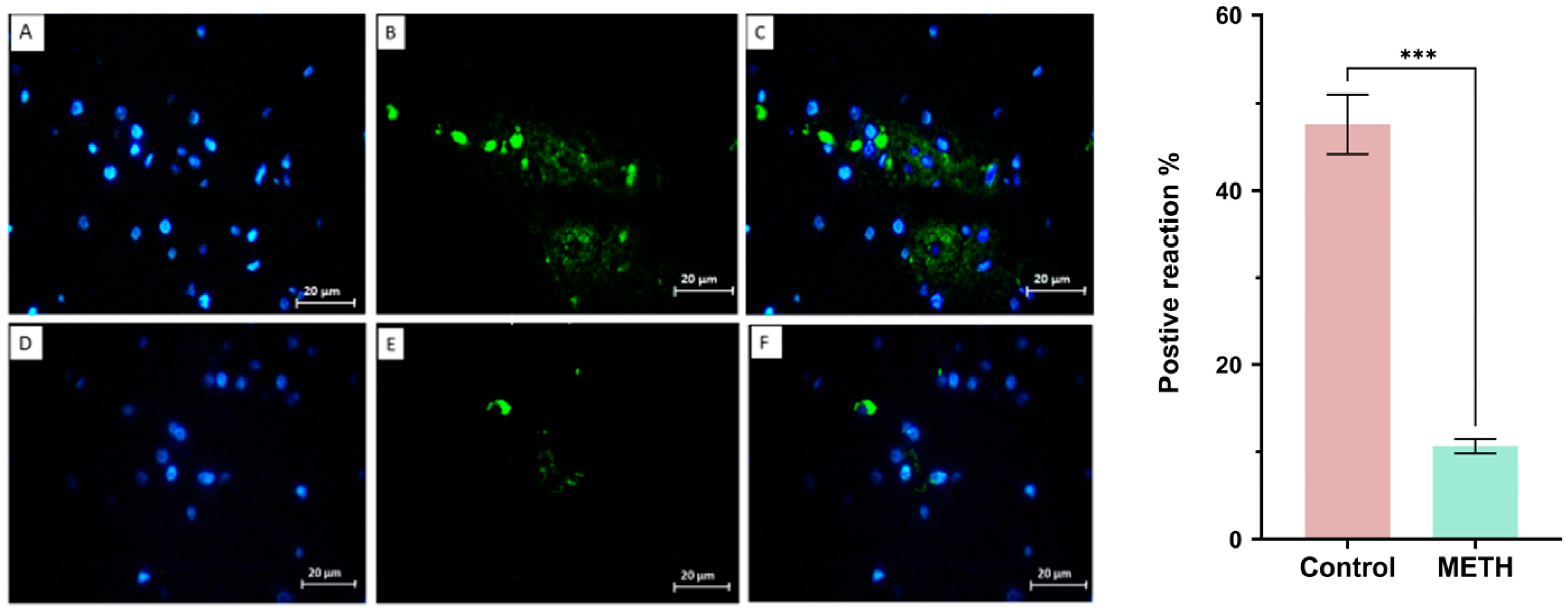
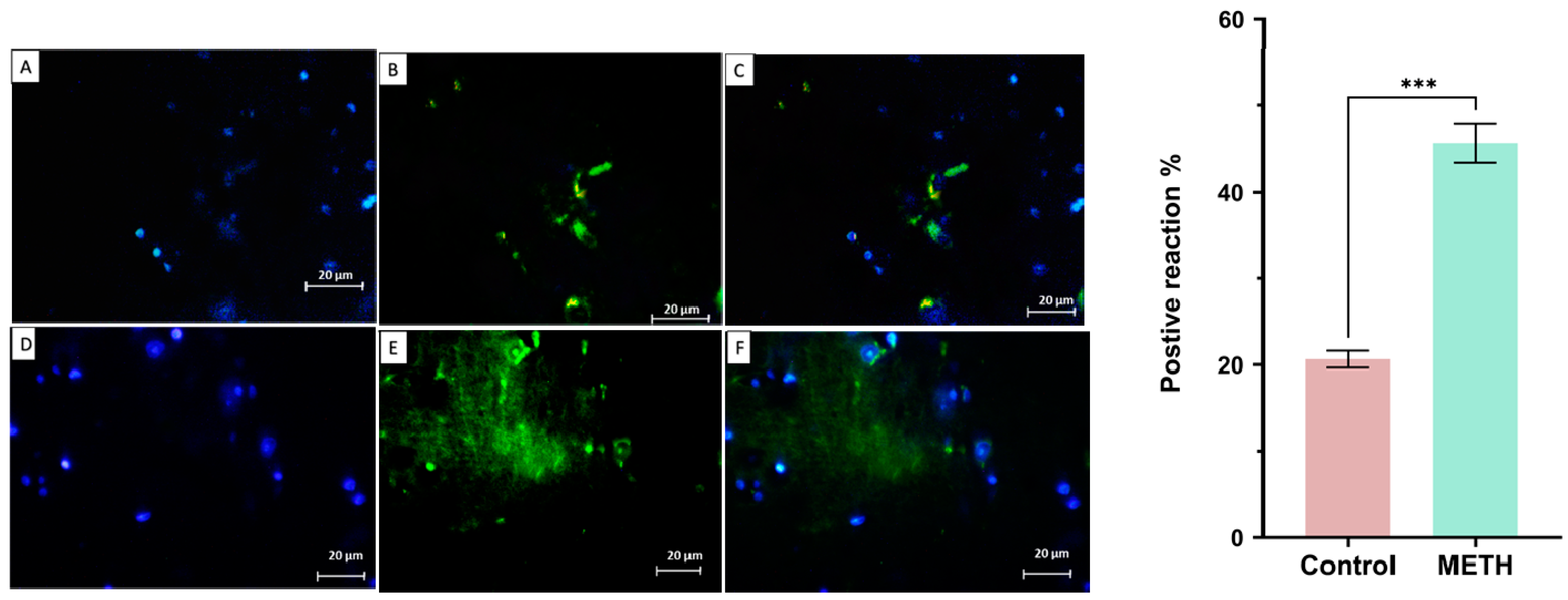
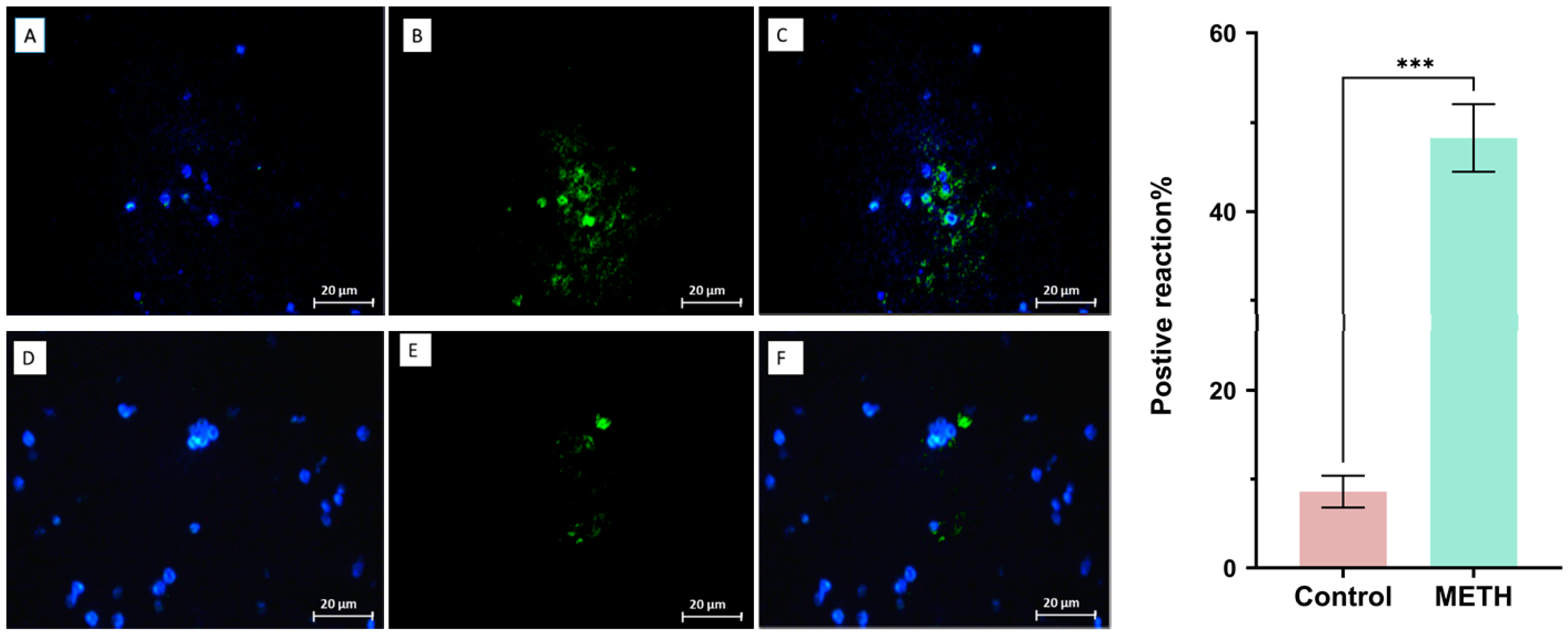
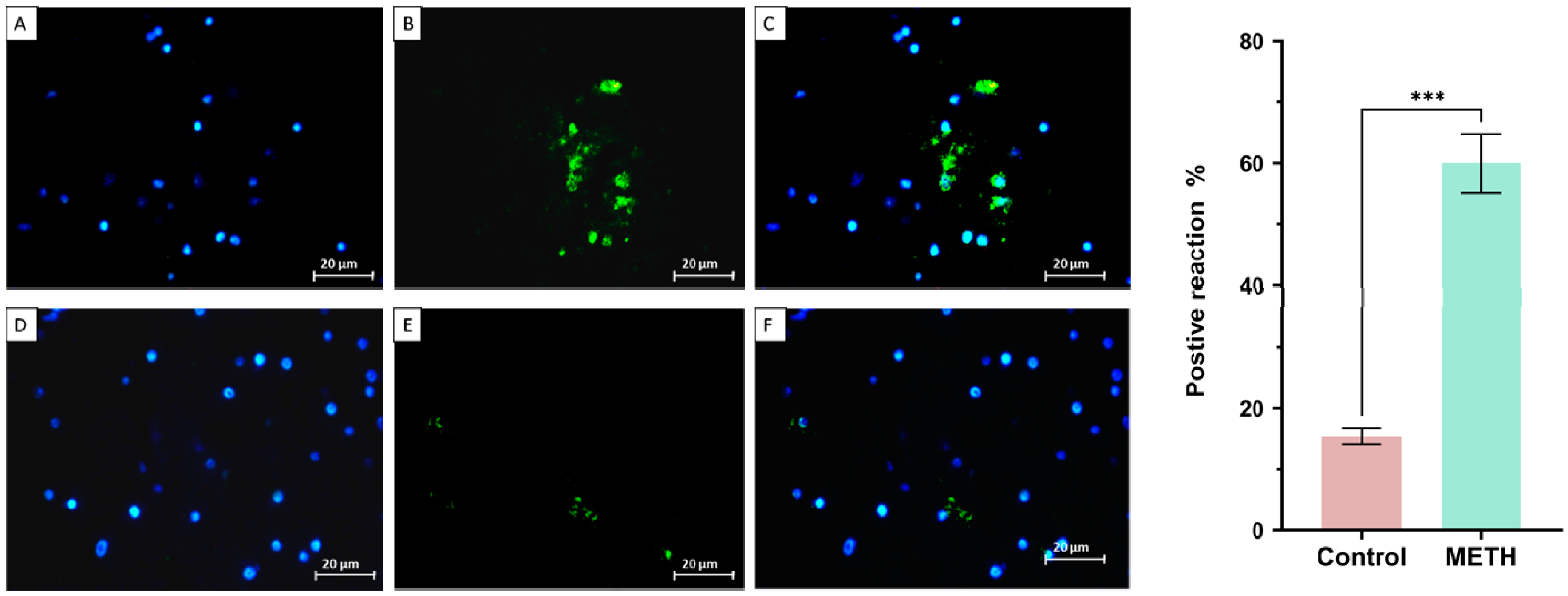
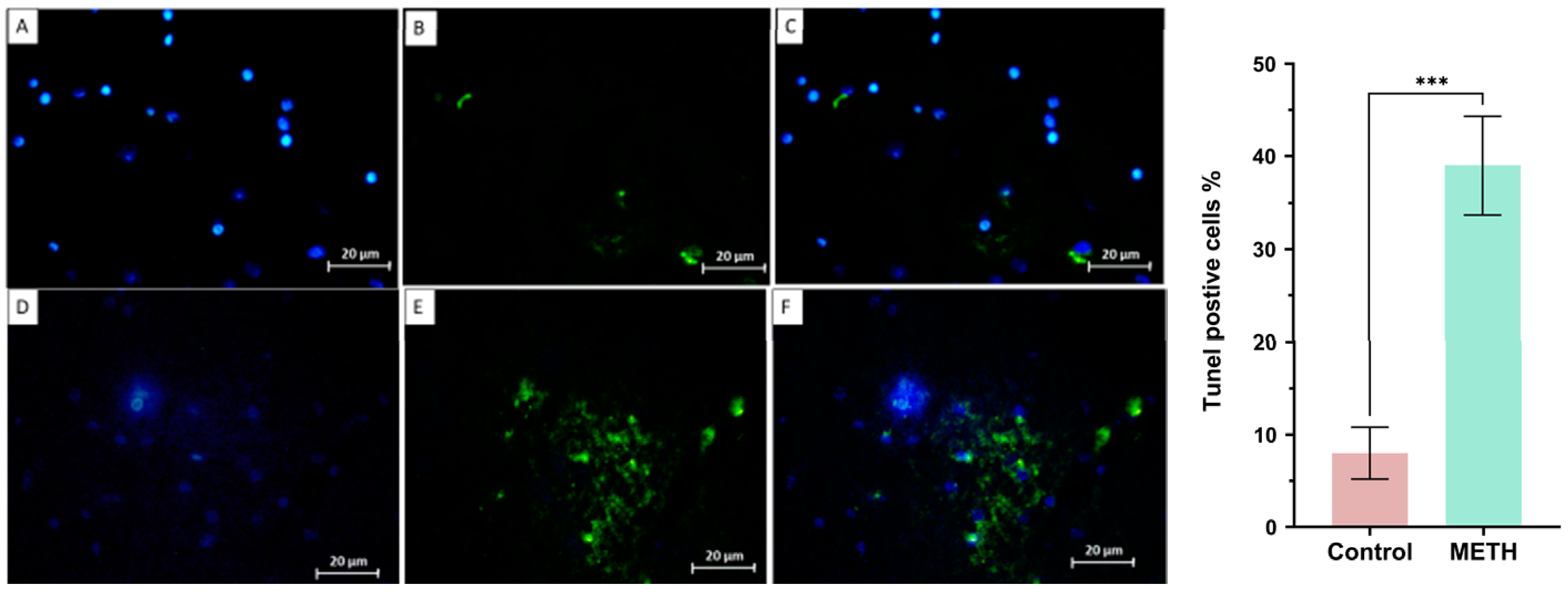
Tunel assay
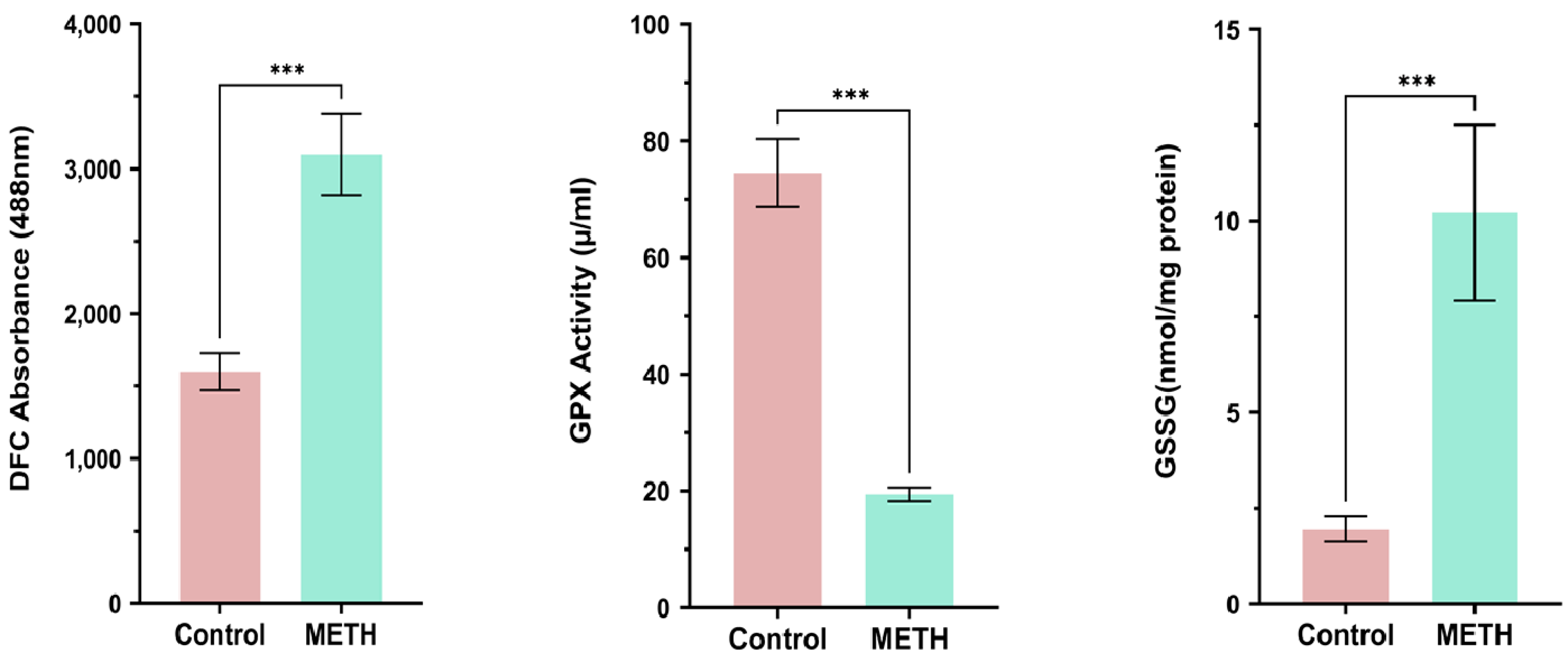
GPX Activation, GSSG, ROS
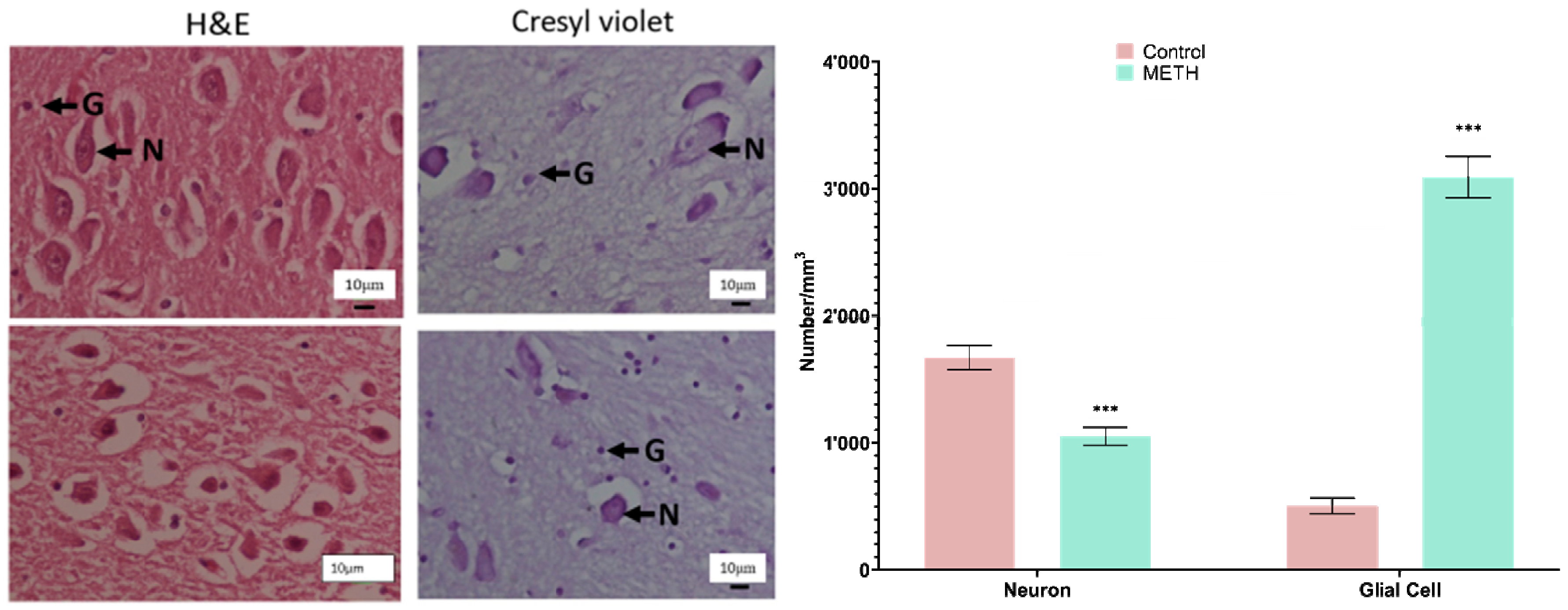
Stereological analysis of the amygdala
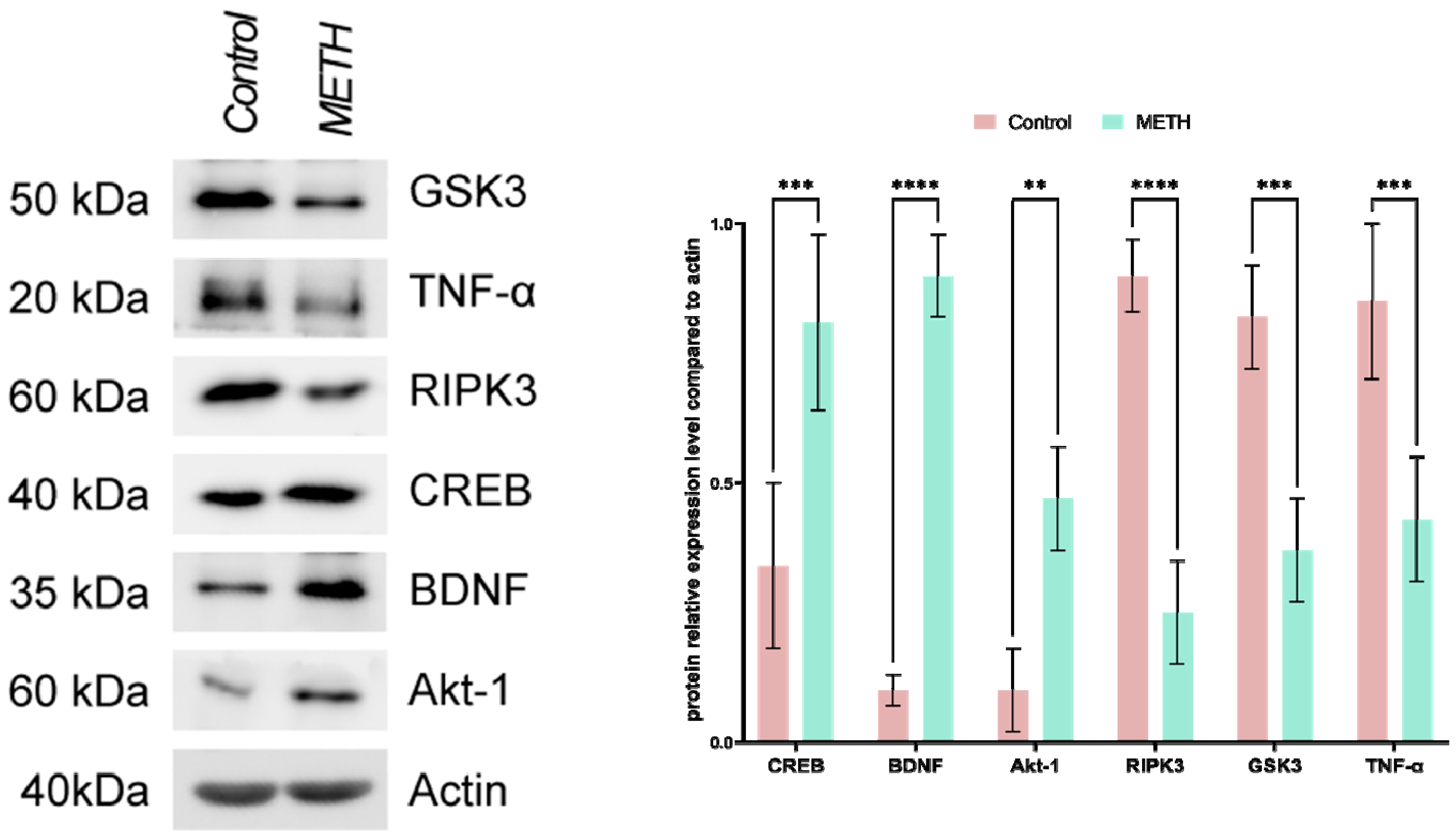
Immunoblotting
3. Discussion
4. Materials and methods
Sample preparation
Tissue Preparation
Real-time PCR
| GENE | FORWARD | REVERSE | Accession No |
|---|---|---|---|
| CREB | CCAGGTGTAGTTTCTCGAGGTGC | TGTTGGTGAGTCTCGAGGC | NM_004379 |
| BDNF | CATCCGAGGACAAGGTGGCTTG | GCCGAACTTTCTGGTCCTCATC | NM_001143810, |
|
Akt-1 GSK3 TNF-α |
TGGACTACCTGCACTCGGAGAA CCGACTAACACCACTGGAAGCT CTCTTCTGCCTGCTGCACTTTG |
GTGCCGCAAAAGGTCTTCATGG AGGATGGTAGCCAGAGGTGGC ATGGGCTACAGGCTTGTCACTC |
NM_005163.2, NM_002093.3 NM_000594 |
| BAX | TCAGGATGCGTCCACCAAGACC | TGTGTCCACGGCGGCAATCACTA | NM_001291428 |
| CASPASE3 | TCAGAGGAGACCGATGCAAGAC | AAGTCGGCCTCCACTGGAATAC | NM_004346 |
| LC3 | TCCAGCACCTTAGTAACCCACG | TGTAGAACGAGGACACTGGCAC | NM_026160 |
| ATG5 | TGCAGATGGACAGTTGCACACG | GCTCAGATGTTCACTCAGCCAC | NM_001286106 |
|
GAPDH RIPK3 BCL2 |
TGCTACAACTGCCACCCAGAAG ATCGCCCTGTGGATGACTGAGT GAAGACACGGCACTCCTTGCA |
ATTGGCAACAGGTACACGGAAG CTTGAGGCAGTAGTTCTTGGTGG GCCAGGAGAAATCAAACAGAGGCC |
NM_001256799 NM_019955.2 NM_000633 |
Immunohistochemistry
H&E and Nissl staining
Stereological study
Number of neurons and glial cells
Tunel assay
Reactive oxygen species (ROS)
Glutathione disulfide content assessments and GSSG assay
SDS-PAGE sample preparation, gel preparation, and running conditions.
Immunoblotting
Statistical analysis
5. Conclusions
Ethical publication statement
Declaration of Competing Interest
Acknowledgment
References
- Behl, T., et al., Current trends in neurodegeneration: Cross talks between oxidative stress, cell death, and inflammation. International Journal of Molecular Sciences, 2021. 22(14): p. 7432. [CrossRef]
- Tahmasebinia, F. and S. Emadi. Effects of clioquinol on the aggregation of beta-amyloid peptides in the presence and absence of metal ions and astrocyte-mediated inflammation. in EUROPEAN JOURNAL OF IMMUNOLOGY. 2016. WILEY-BLACKWELL 111 RIVER ST, HOBOKEN 07030-5774, NJ USA.
- Pourgholaminejad, A. and F. Tahmasebinia, The Role of Th17 Cells in Immunopathogenesis of Neuroinflammatory Disorders. Neuroimmune Diseases: From Cells to the Living Brain, 2019: p. 83.
- Mahmoudiasl, G., Abbaszadeh, H., Rezaei-Tavirani, M., Abdollahifar, M., Khoramgah, M., Niknazar, S., Roozbahany, N. (2019). Nod-like receptor protein 3 and nod-like receptor protein 1 inflammasome activation in the hippocampal region of postmortem METH chronic user. Bratislavske lekarske listy, 120(10), 769-776.
- Khoshsirat, S., Khoramgah, M. S., Mahmoudiasl, G.-R., Rezaei-Tavirani, M., Abdollahifar, M.-A., Tahmasebinia, F., . . . Abbaszadeh, H. A. (2020). LC3 and ATG5 overexpression and neuronal cell death in the prefrontal cortex of postmortem chronic METH users. Journal of Chemical Neuroanatomy, 107, 101802.
- Rasti Boroojeni F, Mashayekhan S, Abbaszadeh HA, Ansarizadeh M, Khoramgah MS, Rahimi Movaghar V. Bioinspired nanofiber scaffold for differentiating bone marrow-derived neural stem cells to oligodendrocyte-like cells: design, fabrication, and characterization. International Journal of Nanomedicine. 2020 Jun 2:3903-20.
- Cadet, J.L. and I.N. Krasnova, Molecular bases of methamphetamine-induced neurodegeneration. International review of neurobiology, 2009. 88: p. 101-119.
- İvecen, B. and O. GOKDEMIR, Methamphetamine Addiction. DAHUDER Medical Journal, 2022. 2(4): p. 98-101.
- Prakash, M.D., et al., Methamphetamine: effects on the brain, gut and immune system. Pharmacological research, 2017. 120: p. 60-67. [CrossRef]
- Chavda, V., et al., Narcolepsy—A Neuropathological Obscure Sleep Disorder: A Narrative Review of Current Literature. Brain Sciences, 2022. 12(11): p. 1473. [CrossRef]
- Rusyniak, D.E., Neurologic manifestations of chronic methamphetamine abuse. Psychiatric Clinics, 2013. 36(2): p. 261-275. [CrossRef]
- Compton, W.M. and N.D. Volkow, Abuse of prescription drugs and the risk of addiction. Drug and alcohol dependence, 2006. 83: p. S4-S7. [CrossRef]
- Downey, L.A. and J.M. Loftis, Altered energy production, lowered antioxidant potential, and inflammatory processes mediate CNS damage associated with abuse of the psychostimulants MDMA and methamphetamine. European journal of pharmacology, 2014. 727: p. 125-129. [CrossRef]
- Varner, K.J., et al., Cardiovascular responses elicited by the “binge” administration of methamphetamine. Journal of Pharmacology and Experimental Therapeutics, 2002. 301(1): p. 152-159. [CrossRef]
- Tehrani, A.M., et al., Methamphetamine induces neurotoxicity-associated pathways and stereological changes in prefrontal cortex. Neuroscience letters, 2019. 712: p. 134478. [CrossRef]
- Volkow, N.D., et al., Association of dopamine transporter reduction with psychomotor impairment in methamphetamine abusers. American Journal of Psychiatry, 2001. 158(3): p. 377-382. [CrossRef]
- Shrestha, P., et al., Methamphetamine induced neurotoxic diseases, molecular mechanism, and current treatment strategies. Biomedicine & Pharmacotherapy, 2022. 154: p. 113591. [CrossRef]
- Hong, S., et al., Expression of dopamine transporter in the different cerebral regions of methamphetamine-dependent rats. Human & Experimental Toxicology, 2015. 34(7): p. 707-717. [CrossRef]
- Yuan, J., et al., Dopamine transporter dysfunction in Han Chinese people with chronic methamphetamine dependence after a short-term abstinence. Psychiatry Research: Neuroimaging, 2014. 221(1): p. 92-96. [CrossRef]
- London, E.D., et al., Mood disturbances and regional cerebral metabolic abnormalities inrecently abstinent methamphetamine abusers. Archives of general psychiatry, 2004. 61(1): p. 73-84. [CrossRef]
- Moszczynska, A., et al., Why is parkinsonism not a feature of human methamphetamine users? Brain, 2004. 127(2): p. 363-370.
- Thompson, P.M., et al., Structural abnormalities in the brains of human subjects who use methamphetamine. Journal of Neuroscience, 2004. 24(26): p. 6028-6036. [CrossRef]
- Miller, D.B. and J.P. O’Callaghan, Elevated environmental temperature and methamphetamine neurotoxicity. Environmental research, 2003. 92(1): p. 48-53. [CrossRef]
- Jayanthi, S., et al., Calcineurin/NFAT-induced up-regulation of the Fas ligand/Fas death pathway is involved in methamphetamine-induced neuronal apoptosis. Proceedings of the National Academy of Sciences, 2005. 102(3): p. 868-873. [CrossRef]
- Kiyatkin, E.A. and H.S. Sharma, Permeability of the blood–brain barrier depends on brain temperature. Neuroscience, 2009. 161(3): p. 926-939. [CrossRef]
- Nopparat, C., et al., The mechanism for the neuroprotective effect of melatonin against methamphetamine-induced autophagy. Journal of pineal research, 2010. 49(4): p. 382-389. [CrossRef]
- Huang, X., et al., Methamphetamine abuse impairs motor cortical plasticity and function. Molecular psychiatry, 2017. 22(9): p. 1274-1281. [CrossRef]
- Salo, R., et al., Behavioral regulation in methamphetamine abusers: an fMRI study. Psychiatry Research: Neuroimaging, 2013. 211(3): p. 234-238. [CrossRef]
- Morales, A.M., et al., Gray-matter volume in methamphetamine dependence: cigarette smoking and changes with abstinence from methamphetamine. Drug and alcohol dependence, 2012. 125(3): p. 230-238. [CrossRef]
- Jernigan, T.L., et al., Effects of methamphetamine dependence and HIV infection on cerebral morphology. American Journal of Psychiatry, 2005. 162(8): p. 1461-1472. [CrossRef]
- Gonçalves, J., et al., Methamphetamine-induced neuroinflammation and neuronal dysfunction in the mice hippocampus: preventive effect of indomethacin. European Journal of Neuroscience, 2010. 31(2): p. 315-326. [CrossRef]
- Khoshsirat, S., et al., LC3 and ATG5 overexpression and neuronal cell death in the prefrontal cortex of postmortem chronic methamphetamine users. Journal of Chemical Neuroanatomy, 2020. 107: p. 101802. [CrossRef]
- Mirakabad, F.S.T., et al., NUPR1-CHOP experssion, autophagosome formation and apoptosis in the postmortem striatum of chronic methamphetamine user. Journal of Chemical Neuroanatomy, 2021. 114: p. 101942. [CrossRef]
- Roohbakhsh, A., K. Shirani, and G. Karimi, Methamphetamine-induced toxicity: The role of autophagy? Chemico-biological interactions, 2016. 260: p. 163-167.
- Mata, M.M., et al., Methamphetamine decreases CD4 T cell frequency and alters pro-inflammatory cytokine production in a model of drug abuse. European journal of pharmacology, 2015. 752: p. 26-33. [CrossRef]
- Mahmoudiasl, G., et al., Nod-like receptor protein 3 and nod-like receptor protein 1 inflammasome activation in the hippocampal region of postmortem methamphetamine chronic user. Bratislavske lekarske listy, 2019. 120(10): p. 769-776. [CrossRef]
- Kohno, M., et al., The relationship between interleukin-6 and functional connectivity in methamphetamine users. Neuroscience letters, 2018. 677: p. 49-54. [CrossRef]
- Guo, D., et al., Molecular mechanisms of programmed cell death in methamphetamine-induced neuronal damage. Frontiers in Pharmacology, 2022. 13. [CrossRef]
- Wu, C.-W., et al., Enhanced oxidative stress and aberrant mitochondrial biogenesis in human neuroblastoma SH-SY5Y cells during methamphetamine induced apoptosis. Toxicology and applied pharmacology, 2007. 220(3): p. 243-251. [CrossRef]
- Gholami, M., et al., Pharmacological and molecular evidence of neuroprotective curcumin effects against biochemical and behavioral sequels caused by methamphetamine: Possible function of CREB-BDNF signaling pathway. Basic and Clinical Neuroscience, 2021. 12(3): p. 325. [CrossRef]
- Motaghinejad, M., et al., Possible involvement of CREB/BDNF signaling pathway in neuroprotective effects of topiramate against methylphenidate induced apoptosis, oxidative stress and inflammation in isolated hippocampus of rats: molecular, biochemical and histological evidences. Brain research bulletin, 2017. 132: p. 82-98. [CrossRef]
- Motaghinejad, M., et al., The possible role of CREB-BDNF signaling pathway in neuroprotective effects of minocycline against alcohol-induced neurodegeneration: molecular and behavioral evidences. Fundamental & Clinical Pharmacology, 2021. 35(1): p. 113-130. [CrossRef]
- Feizipour, S., et al., Selegiline acts as neuroprotective agent against methamphetamine-prompted mood and cognitive related behavior and neurotoxicity in rats: Involvement of CREB/BDNF and Akt/GSK3 signal pathways. Iranian Journal of Basic Medical Sciences, 2020. 23(5): p. 606. [CrossRef]
- Keshavarzi, S., et al., Protective role of metformin against methamphetamine induced anxiety, depression, cognition impairment and neurodegeneration in rat: the role of CREB/BDNF and Akt/GSK3 signaling pathways. Neurotoxicology, 2019. 72: p. 74-84. [CrossRef]
- Darabi, S., et al., SMER28 attenuates dopaminergic toxicity mediated by 6-hydroxydopamine in the rats via modulating oxidative burdens and autophagy-related parameters. Neurochemical Research, 2018. 43: p. 2313-2323. [CrossRef]
- Darabi, S., et al., Creatine and retinoic acid effects on the induction of autophagy and differentiation of adipose tissue-derived stem cells into GABAergic-like neurons. Journal of Babol university of medical sciences, 2017. 19(8): p. 41-49.
- Abbaszadeh, H.-A., et al., Bone marrow stromal cell transdifferentiation into oligodendrocyte-like cells using triiodothyronine as a inducer with expression of platelet-derived growth factor α as a maturity marker. Iranian biomedical journal, 2013. 17(2): p. 62. [CrossRef]
- Moghimi, N., et al., COVID-19 disrupts spermatogenesis through the oxidative stress pathway following induction of apoptosis. Apoptosis, 2021. 26(7-8): p. 415-430. [CrossRef]
- Shaerzadeh, F., et al., Methamphetamine neurotoxicity, microglia, and neuroinflammation. Journal of neuroinflammation, 2018. 15: p. 1-6.
- Castellano, P., et al., Methamphetamine compromises gap junctional communication in astrocytes and neurons. Journal of neurochemistry, 2016. 137(4): p. 561-575. [CrossRef]
- Mayr, B. and M. Montminy, Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature reviews Molecular cell biology, 2001. 2(8): p. 599-609. [CrossRef]
- Yang, G., et al., Ginsenoside Rb1 attenuates methamphetamine (METH)-induced neurotoxicity through the NR2B/ERK/CREB/BDNF signalings in vitro and in vivo models. Journal of Ginseng Research, 2022. 46(3): p. 426-434. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




