Submitted:
25 May 2023
Posted:
26 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
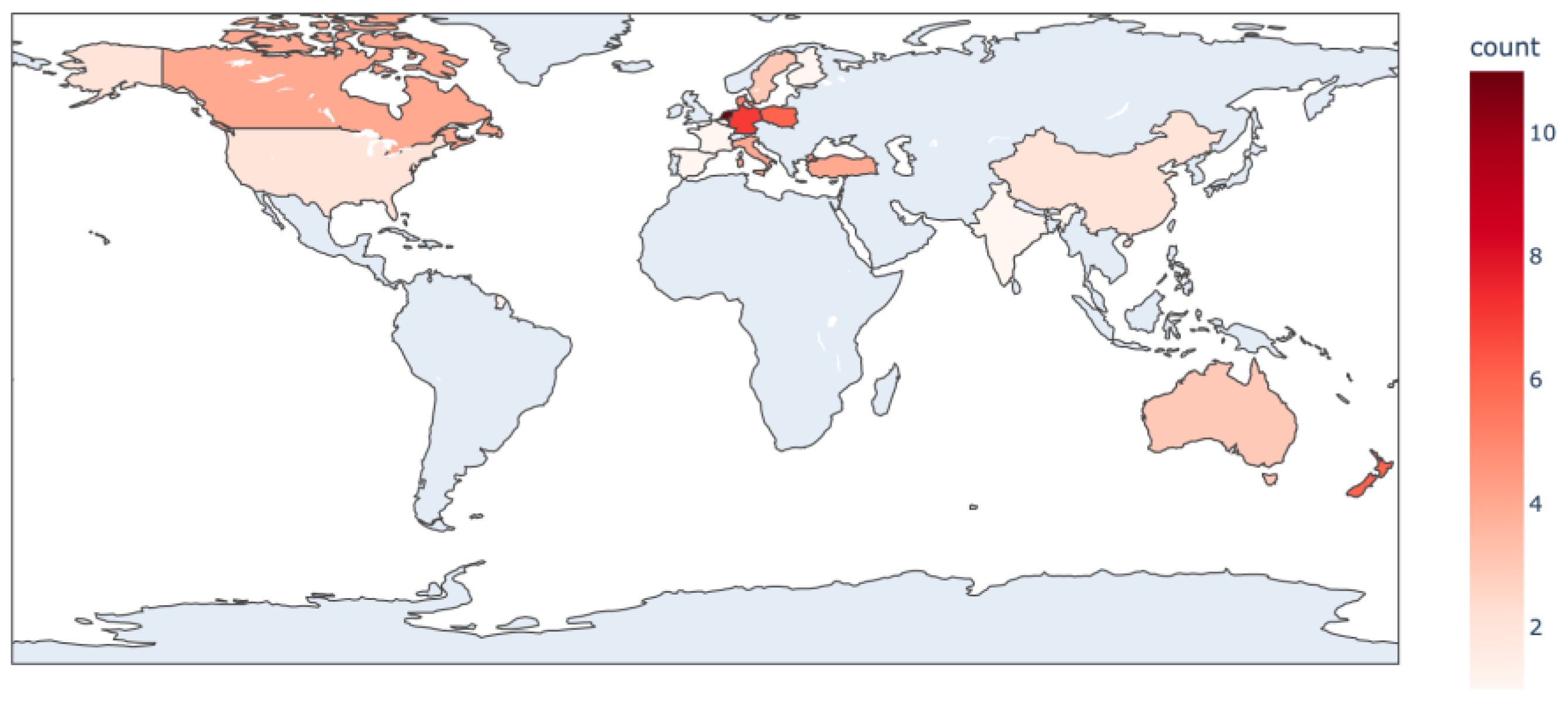
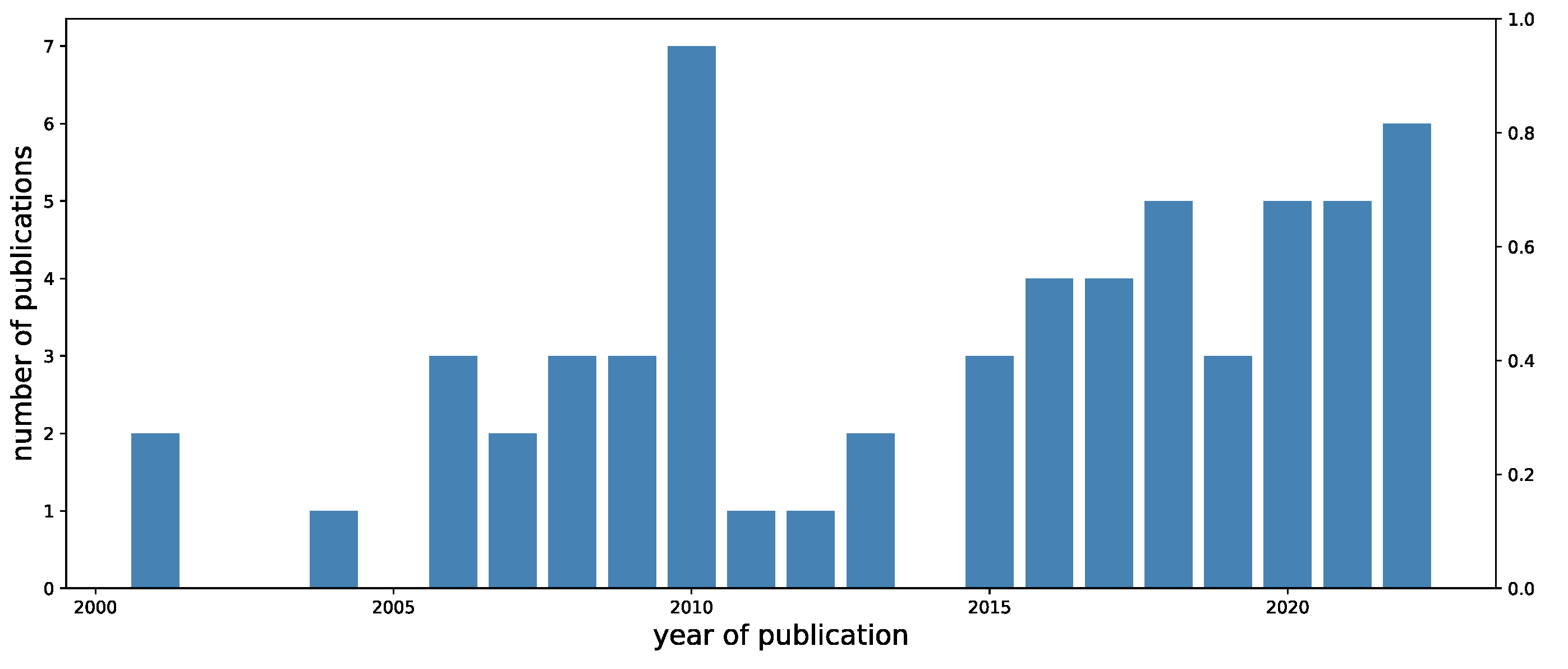
2. Review Methodology and Results
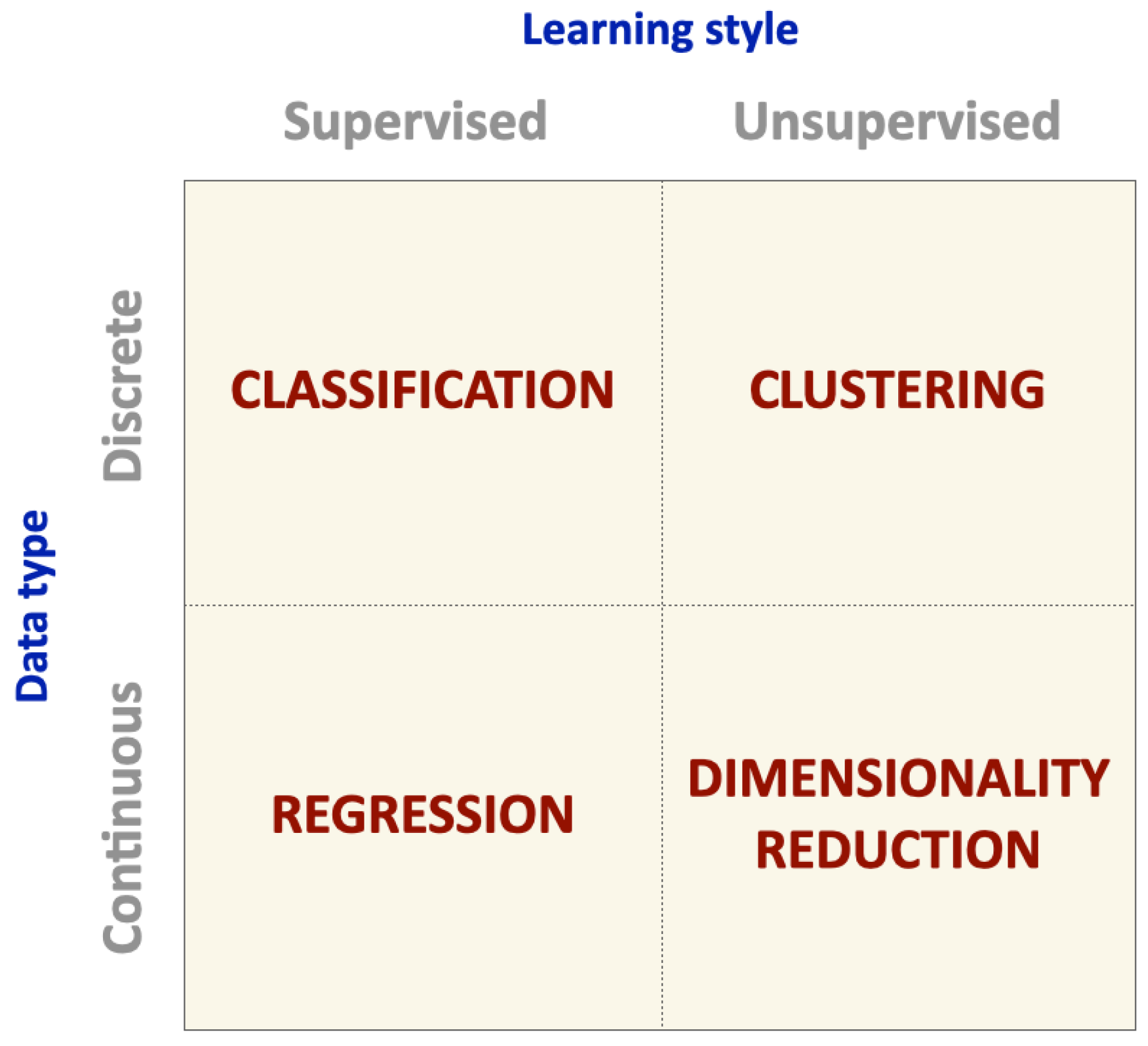
3. Machine Learning Algorithms and Performance Metrics
4. Application of Modeling Approaches in AMS
4.1. Health
4.1.1. Mastitis
4.1.2. Other Diseases
4.2. Cows Behaviour and Hard Management
- a fully CNN that detects the landmarks in the image;
- a CNN that works with the probability map produced by the first CNN as input to detect the cows and their orientations.
4.3. Production
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vijayakumar, M.; Park, J.H.; Ki, K.S.; Lim, D.H.; Kim, S.B.; Park, S.M.; Jeong, H.Y.; Park, B.Y.; Kim, T.I. The effect of lactation number, stage, length, and milking frequency on milk yield in Korean Holstein dairy cows using automatic milking system. Asian-Australasian J. Anim. Sci. 2017, 30, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Tse, C.; Barkema, H.W.; DeVries, T.J.; Rushen, J.; Pajor, E.A. Impact of automatic milking systems on dairy cattle producers’ reports of milking labour management, milk production and milk quality. Animal 2018, 12, 2649–2656. [Google Scholar] [CrossRef] [PubMed]
- Österman, S.; Redbo, I. Effects of milking frequency on lying down and getting up behaviour in dairy cows. Appl. Anim. Behav. Sci. 2000, 70, 167–176. [Google Scholar] [CrossRef]
- Jacobs, J.A.; Siegford, J.M. Invited review: The impact of automatic milking systems on dairy cow management, behavior, health, and welfare. J. Dairy Sci. 2012, 95, 2227–2247. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Pacheco, G.G.; Kaler, J.; Remnant, J.; Cheyne, L.; Abbott, C.; French, A.P.; Pridmore, T.P.; Huxley, J.N. Behavioural changes in dairy cows with lameness in an automatic milking system. Appl. Anim. Behav. Sci. 2014, 150, 1–8. [Google Scholar] [CrossRef]
- Gargiulo, J.; Lyons, N.; Kempton, K.; Armstrong, D.; Garcia, S. Physical and economic comparison of pasture-based automatic and conventional milking systems. J. Dairy Sci. 2020, 103, 8231–8240. [Google Scholar] [CrossRef] [PubMed]
- Bonora, F.; Benni, S.; Barbaresi, A.; Tassinari, P.; Torreggiani, D. A cluster-graph model for herd characterisation in dairy farms equipped with an automatic milking system. Biosyst. Eng. 2018, 167, 1–7. [Google Scholar] [CrossRef]
- Wolfert, S.; Ge, L.; Verdouw, C.; Bogaardt, M.-J. Big Data in Smart Farming – A review. Agric. Syst. 2017, 153, 69–80. [Google Scholar] [CrossRef]
- Cockburn, M. Review: Application and Prospective Discussion of Machine Learning for the Management of Dairy Farms. Animals 2020, 10, 1690. [Google Scholar] [CrossRef]
- Nayeri, S.; Sargolzaei, M.; Tulpan, D. A review of traditional and machine learning methods applied to animal breeding. Anim. Heal. Res. Rev. 2019, 20, 31–46. [Google Scholar] [CrossRef]
- Rebala, G.; Ravi, A.; Churiwala, S. Machine Learning Definition and Basics. In An Introduction to Machine Learning; Springer: Cham, 2019; pp. 1–17. [Google Scholar]
- García, R.; Aguilar, J.; Toro, M.; Pinto, A.; Rodríguez, P. A systematic literature review on the use of machine learning in precision livestock farming. Comput. Electron. Agric. 2020, 179, 105826. [Google Scholar] [CrossRef]
- Wang, Z.; Shadpour, S.; Chan, E.; Rotondo, V.; Wood, K.M.; Tulpan, D. ASAS-NANP SYMPOSIUM: Applications of machine learning for livestock body weight prediction from digital images. J. Anim. Sci. 2021, 99, skab022. [Google Scholar] [CrossRef] [PubMed]
- Brock, J.; Lange, M.; Tratalos, J.A.; More, S.J.; Graham, D.A.; Guelbenzu-Gonzalo, M.; Thulke, H.-H. Combining expert knowledge and machine-learning to classify herd types in livestock systems. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Jiang, M.; Wang, W.; Zhang, W.; Wang, R. On-farm welfare monitoring system for goats based on Internet of Things and machine learning. Int. J. Distrib. Sens. Networks 2020, 16. [Google Scholar] [CrossRef]
- Carslake, C.; Vázquez-Diosdado, J.A.; Kaler, J. Machine Learning Algorithms to Classify and Quantify Multiple Behaviours in Dairy Calves Using a Sensor: Moving beyond Classification in Precision Livestock. Sensors 2020, 21, 88. [Google Scholar] [CrossRef]
- Higaki, S.; Miura, R.; Suda, T.; Andersson, L.M.; Okada, H.; Zhang, Y.; Itoh, T.; Miwakeichi, F.; Yoshioka, K. Estrous detection by continuous measurements of vaginal temperature and conductivity with supervised machine learning in cattle. Theriogenology 2018, 123, 90–99. [Google Scholar] [CrossRef]
- Bovo, M.; Agrusti, M.; Benni, S.; Torreggiani, D.; Tassinari, P. Random Forest Modelling of Milk Yield of Dairy Cows under Heat Stress Conditions. Animals 2021, 11, 1305. [Google Scholar] [CrossRef]
- Gorczyca, M.T.; Gebremedhin, K.G. Ranking of environmental heat stressors for dairy cows using machine learning algorithms. Comput. Electron. Agric. 2019, 168, 105124. [Google Scholar] [CrossRef]
- Ren, K.; Bernes, G.; Hetta, M.; Karlsson, J. Tracking and analysing social interactions in dairy cattle with real-time locating system and machine learning. J. Syst. Arch. 2021, 116. [Google Scholar] [CrossRef]
- Lokhorst, C.; De Mol, R.M.; Kamphuis, C. Invited Review: Big Data in Precision Dairy Farming. Animal 2019, 13, 1519. [Google Scholar] [CrossRef]
- Slob, N.; Catal, C.; Kassahun, A. Application of machine learning to improve dairy farm management: A systematic literature review. Prev. Veter- Med. 2020, 187, 105237. [Google Scholar] [CrossRef]
- Samuel, A. L. Some studies in machine learning using the game of checkers, in IBM Journal of Research and Development, vol. 44, no. 1.2, 2000. 206; 226. [Google Scholar]
- Sharma, R.; Kamble, S.S.; Gunasekaran, A.; Kumar, V.; Kumar, A. A systematic literature review on machine learning applications for sustainable agriculture supply chain performance. Comput. Oper. Res. 2020, 119, 104926. [Google Scholar] [CrossRef]
- Sharma, D.; Kumar, N. A Review on Machine Learning Algorithms, Tasks and Applications. Int. J. Adv. Res. 2017, 6, 1548–1552. [Google Scholar]
- Tarca, A.L.; Carey, V.J.; Chen, X.-W.; Romero, R.; Drăghici, S. Machine Learning and Its Applications to Biology. PLOS Comput. Biol. 2007, 3, e116. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Kaelbling, L.P.; Littman, M.L.; Moore, A.W. Reinforcement Learning: A Survey. J. Artif. Intell. Res. 1996, 4, 237–285. [Google Scholar] [CrossRef]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge: Cambridge University Press, 2014. [Google Scholar] [CrossRef]
- Ramsey, F.; Schafer, D. The Statistical Sleuth: A Course in Methods of Data Analysis. 1st ed. Wadsworth Publishing Company, 1996, Belmont CA. 1996. [Google Scholar]
- Kingsford, C.; Salzberg, S.L. What are decision trees? Nat. Biotechnol. 2008, 26, 1011–1013. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Cielen, D.; Meysman, A.D.B.; Ali, M. Introducing Data Science: Big Data, Machine Learning, and More, Using Python Tools. 2016, New York: Manning Publications Co.
- Vapnik, V.; Izmailov, R. Knowledge transfer in SVM and neural networks. Ann. Math. Artif. Intell. 2017, 81, 3–19. [Google Scholar] [CrossRef]
- Jensen, F.V.; Nielsen, T.D. Bayesian networks and decision graphs (Vol. 2). 2007, New York: Springer.
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy sets. Inf. Control 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Goldberg, D.E. Genetic Algorithms in Search Optimization and Machine Learning. 1989, Addison-Wesley, Reading, MA.
- Shapiro, J. Genetic algorithms in machine learning. In: Advanced Course on Artificial Intelligence, 1999 Springer, Berlin, Heidelberg, pp. 146-168.
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine Learning in Agriculture: A Comprehensive Updated Review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef]
- Melchior, M.; Vaarkamp, H.; Fink-Gremmels, J. Biofilms: A role in recurrent mastitis infections? Veter- J. 2006, 171, 398–407. [Google Scholar] [CrossRef]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. A large-scale study of indicators of sub-clinical mastitis in dairy cattle by attribute weighting analysis of milk composition features: highlighting the predictive power of lactose and electrical conductivity. J. Dairy Res. 2018, 85, 193–200. [Google Scholar] [CrossRef]
- Cavero, D.; Tölle, K.-H.; Buxadé, C.; Krieter, J. Mastitis detection in dairy cows by application of fuzzy logic. Livest. Sci. 2006, 105, 207–213. [Google Scholar] [CrossRef]
- Steeneveld, W.; van der Gaag, L.; Ouweltjes, W.; Mollenhorst, H.; Hogeveen, H. Discriminating between true-positive and false-positive clinical mastitis alerts from automatic milking systems. J. Dairy Sci. 2010, 93, 2559–2568. [Google Scholar] [CrossRef]
- Sharma, N.; Singh, N.K.; Bhadwal, M.S. Relationship of Somatic Cell Count and Mastitis: An Overview. Asian-Australasian J. Anim. Sci. 2011, 24, 429–438. [Google Scholar] [CrossRef]
- Zecconi, A.; Dell’orco, F.; Vairani, D.; Rizzi, N.; Cipolla, M.; Zanini, L. Differential Somatic Cell Count as a Marker for Changes of Milk Composition in Cows with Very Low Somatic Cell Count. Animals 2020, 10, 604. [Google Scholar] [CrossRef]
- Bobbo, T.; Biffani, S.; Taccioli, C.; Penasa, M.; Cassandro, M. Comparison of machine learning methods to predict udder health status based on somatic cell counts in dairy cows. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. Hierarchical pattern recognition in milking parameters predicts mastitis prevalence. Comput. Electron. Agric. 2018, 147, 6–11. [Google Scholar] [CrossRef]
- Norberg, E.; Hogeveen, H.; Korsgaard, I.; Friggens, N.; Sloth, K.; Løvendahl, P. Electrical Conductivity of Milk: Ability to Predict Mastitis Status. J. Dairy Sci. 2004, 87, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, C.; Sherlock, R.; Jago, J.; Mein, G.; Hogeveen, H. Automatic Detection of Clinical Mastitis Is Improved by In-Line Monitoring of Somatic Cell Count. J. Dairy Sci. 2008, 91, 4560–4570. [Google Scholar] [CrossRef] [PubMed]
- Mollenhorst, H.; van der Tol, P.; Hogeveen, H. Somatic cell count assessment at the quarter or cow milking level. J. Dairy Sci. 2010, 93, 3358–3364. [Google Scholar] [CrossRef]
- Khatun, M.; Clark, C.E.F.; Lyons, N.A.; Thomson, P.; Kerrisk, K.L.; García, S.C. Early detection of clinical mastitis from electrical conductivity data in an automatic milking system. Anim. Prod. Sci. 2017, 57, 1226–1232. [Google Scholar] [CrossRef]
- Sun, Z.; Samarasinghe, S.; Jago, J. Detection of mastitis and its stage of progression by automatic milking systems using artificial neural networks. J. Dairy Res. 2009, 77, 168–175. [Google Scholar] [CrossRef]
- Hovinen, M.; Aisla, A.-M.; Pyörälä, S. Accuracy and reliability of mastitis detection with electrical conductivity and milk colour measurement in automatic milking. Acta Agric. Scand. Sect. A — Anim. Sci. 2006, 56, 121–127. [Google Scholar] [CrossRef]
- Kamphuis, C.; Pietersma, D.; van der Tol, R.; Wiedemann, M.; Hogeveen, H. Using sensor data patterns from an automatic milking system to develop predictive variables for classifying clinical mastitis and abnormal milk. Comput. Electron. Agric. 2008, 62, 169–181. [Google Scholar] [CrossRef]
- Altay, Y.; Kılıç, B.; Aytekin, I.; Keskin, I. Determination of Factors Affecting Mastitis in Holstein Friesian and Brown Swiss by Using Logistic Regression Analysis. Selcuk J. Agric. Food Sci. 2019, 33, 194–197. [Google Scholar] [CrossRef]
- Chagunda, M.; Friggens, N.; Rasmussen; Larsen, T. A Model for Detection of Individual Cow Mastitis Based on an Indicator Measured in Milk. J. Dairy Sci. 2006, 89, 2980–2998. [Google Scholar] [CrossRef]
- Friggens, N.; Chagunda, M.; Bjerring, M.; Ridder, C.; Hojsgaard, S.; Larsen, T. Estimating Degree of Mastitis from Time-Series Measurements in Milk: A Test of a Model Based on Lactate Dehydrogenase Measurements. J. Dairy Sci. 2007, 90, 5415–5427. [Google Scholar] [CrossRef]
- Ankinakatte, S.; Norberg, E.; Løvendahl, P.; Edwards, D.; Højsgaard, S. Predicting mastitis in dairy cows using neural networks and generalized additive models: A comparison. Comput. Electron. Agric. 2013, 99, 1–6. [Google Scholar] [CrossRef]
- Zank, W.; Schlatterer, B. Assessment of subacute mammary inflammation by soluble biomarkers in comparison to somatic cell counts in quarter milk samples from dairy cows. J. Veter- Med. Ser. A 1998, 45, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Nyman, A.-K.; Emanuelson, U.; Waller, K. Diagnostic test performance of somatic cell count, lactate dehydrogenase, and N-acetyl-β-d-glucosaminidase for detecting dairy cows with intramammary infection. J. Dairy Sci. 2016, 99, 1440–1448. [Google Scholar] [CrossRef]
- Penry, J.; Crump, P.; Ruegg, P.; Reinemann, D. Short communication: Cow- and quarter-level milking indicators and their associations with clinical mastitis in an automatic milking system. J. Dairy Sci. 2017, 100, 9267–9272. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.A.; King, M.T.; Matson, R.D.; DeVries, T.J.; Deardon, R.; Barkema, H.W. Mastitis detection with recurrent neural networks in farms using automated milking systems. Comput. Electron. Agric. 2021, 192, 106618. [Google Scholar] [CrossRef]
- Naqvi, S.A.; King, M.T.; DeVries, T.J.; Barkema, H.W.; Deardon, R. Data considerations for developing deep learning models for dairy applications: A simulation study on mastitis detection. Comput. Electron. Agric. 2022, 196. [Google Scholar] [CrossRef]
- Bonestroo, J.; van der Voort, M.; Hogeveen, H.; Emanuelson, U.; Klaas, I.C.; Fall, N. Forecasting chronic mastitis using automatic milking system sensor data and gradient-boosting classifiers. Comput. Electron. Agric. 2022, 198. [Google Scholar] [CrossRef]
- Mollenhorst, H.; Rijkaart, L.; Hogeveen, H. Mastitis alert preferences of farmers milking with automatic milking systems. J. Dairy Sci. 2012, 95, 2523–2530. [Google Scholar] [CrossRef]
- de Mol, R.; Ouweltjes, W. Detection model for mastitis in cows milked in an automatic milking system. Prev. Veter- Med. 2001, 49, 71–82. [Google Scholar] [CrossRef]
- De Mol, R.; Woldt, W.; Blokhuis, H. Application of Fuzzy Logic in Automated Cow Status Monitoring. J. Dairy Sci. 2001, 84, 400–410. [Google Scholar] [CrossRef]
- Khamaysa Hajaya, M. , S., Kulasiri, G. D., & Lopez Benavides, M. Detection of dairy cattle Mastitis: Modelling of milking features using deep neural networks. In Modelling and Simulation Society of Australia and New Zealand (Eds.), Proceedings of the 23rd International Congress on Modelling and Simulation, 2019, (pp.; pp. 1656–1662.
- Steeneveld, W.; van der Gaag, L.; Barkema, H.; Hogeveen, H. Simplify the interpretation of alert lists for clinical mastitis in automatic milking systems. Comput. Electron. Agric. 2010, 71, 50–56. [Google Scholar] [CrossRef]
- Kamphuis, C.; Mollenhorst, H.; Heesterbeek, J.A.P.; Hogeveen, H. Data Mining to Detect Clinical Mastitis with Automatic Milking. In Proceedings of the 5th IDF Mastitis Conference: Mastitis Research into Practice, Christchurch, New Zealand, 2010a pp. 568-572. 21-24. [Google Scholar]
- Kamphuis, C.; Mollenhorst, H.; Heesterbeek, J.; Hogeveen, H. Detection of clinical mastitis with sensor data from automatic milking systems is improved by using decision-tree induction. J. Dairy Sci. 2010, 93, 3616–3627. [Google Scholar] [CrossRef] [PubMed]
- Bausewein, M.; Mansfeld, R.; Doherr, M.G.; Harms, J.; Sorge, U.S. Sensitivity and Specificity for the Detection of Clinical Mastitis by Automatic Milking Systems in Bavarian Dairy Herds. Animals 2022, 12, 2131. [Google Scholar] [CrossRef] [PubMed]
- Højsgaard, S.; Friggens, N. Quantifying degree of mastitis from common trends in a panel of indicators for mastitis in dairy cows. J. Dairy Sci. 2010, 93, 582–592. [Google Scholar] [CrossRef]
- Sørensen, L.; Bjerring, M.; Løvendahl, P. Monitoring individual cow udder health in automated milking systems using online somatic cell counts. J. Dairy Sci. 2016, 99, 608–620. [Google Scholar] [CrossRef]
- Cavero, D.; Tölle, K.-H.; Henze, C.; Buxadé, C.; Krieter, J. Mastitis detection in dairy cows by application of neural networks. Livest. Sci. 2008, 114, 280–286. [Google Scholar] [CrossRef]
- Krieter, J.; Cavero, D.; Henze, C. Mastitis detection in dairy cows using neural networks. Conference: Agrarinformatik im Spannungsfeld zwischen Regionalisierung und globalen Wertschöpfungsketten–Referate der 27. 2007, GIL Jahrestagung.
- Mammadova, N.; Keskin, I. Application of the Support Vector Machine to Predict Subclinical Mastitis in Dairy Cattle. Sci. World J. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Mammadova, N.M.; Keskin, I. Application of neural network and adaptive neuro-fuzzy inference system to predict subclinical mastitis in dairy cattle. Indian J. Anim. Res. 2015, 49. [Google Scholar] [CrossRef]
- Mammadova, N.M.; Keskin, I. Subclinical mastitis prediction in dairy cattle by application of Fuzzy Logic. Pak. J. Agric. Sci., 2015b, 52(4).
- Anglart, D.; Hallén-Sandgren, C.; Emanuelson, U.; Rönnegård, L. Comparison of methods for predicting cow composite somatic cell counts. J. Dairy Sci. 2020, 103, 8433–8442. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive analysis of machine learning models for prediction of sub-clinical mastitis: Deep Learning and Gradient-Boosted Trees outperform other models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef]
- Tian, F.; Wang, Z.; Yu, S.; Xiong, B.; Wang, S. Clinical mastitis detection by on-line measurements of milk yield, electrical conductivity and deep Learn. In Journal of Physics: Conference Series, 2020, Vol. 1635, No. 1, p. 012046. IOP Publishing. [CrossRef]
- Ghafoor, N.A.; Sitkowska, B. MasPA: A Machine Learning Application to Predict Risk of Mastitis in Cattle from AMS Sensor Data. Agriengineering 2021, 3, 575–583. [Google Scholar] [CrossRef]
- Kamphuis, C.; Mollenhorst, H.; Hogeveen, H. Sensor measurements revealed: Predicting the Gram-status of clinical mastitis causal pathogens. Comput. Electron. Agric. 2011, 77, 86–94. [Google Scholar] [CrossRef]
- Hassan, K.; Samarasinghe, S.; Lopez-Benavides, M. Use of neural networks to detect minor and major pathogens that cause bovine mastitis. J. Dairy Sci. 2009, 92, 1493–1499. [Google Scholar] [CrossRef] [PubMed]
- Horpiencharoen, W.; Thongratsakul, S.; Poolkhet, C. Risk factors of clinical mastitis and antimicrobial susceptibility test results of mastitis milk from dairy cattle in western Thailand: Bayesian network analysis. Prev. Veter- Med. 2019, 164, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Steeneveld, W.; van der Gaag, L.; Barkema, H.; Hogeveen, H. Providing probability distributions for the causal pathogen of clinical mastitis using naive Bayesian networks. J. Dairy Sci. 2009, 92, 2598–2609. [Google Scholar] [CrossRef] [PubMed]
- Castro, Ángel; Pereira, J. M.; Amiama, C.; Bueno, J. Mastitis diagnosis in ten Galician dairy herds (NW Spain) with automatic milking systems. Span. J. Agric. Res. 2015, 13, e0504. [Google Scholar] [CrossRef]
- Liberati, P.; Zappavigna, P. Improving the automated monitoring of dairy cows by integrating various data acquisition systems. Comput. Electron. Agric. 2009, 68, 62–67. [Google Scholar] [CrossRef]
- Steensels, M.; Antler, A.; Bahr, C.; Berckmans, D.; Maltz, E.; Halachmi, I. A decision-tree model to detect post-calving diseases based on rumination, activity, milk yield, BW and voluntary visits to the milking robot. Animal 2016, 10, 1493–1500. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, C.; Wang, H.; Xu, W.; Zhao, Z.; Chen, M.; Jia, B.; Huang, B. The Early Prediction of Common Disorders in Dairy Cows Monitored by Automatic Systems with Machine Learning Algorithms. Animals 2022, 12, 1251. [Google Scholar] [CrossRef]
- Scott, V.; Thomson, P.; Kerrisk, K.; Garcia, S. Influence of provision of concentrate at milking on voluntary cow traffic in a pasture-based automatic milking system. J. Dairy Sci. 2014, 97, 1481–1490. [Google Scholar] [CrossRef]
- Adamczyk, K.; Cywicka, D.; Herbut, P.; Trześniowska, E. The application of cluster analysis methods in assessment of daily physical activity of dairy cows milked in the Voluntary Milking System. Comput. Electron. Agric. 2017, 141, 65–72. [Google Scholar] [CrossRef]
- Banhazi, T.M.; Tscharke, M. A brief review of the application of machine vision in livestock behaviour analysis. J. Agric. Informatics 2016, 7, 23–42. [Google Scholar] [CrossRef]
- Guzhva, O.; Ardö, H.; Herlin, A.; Nilsson, M.; Åström, K.; Bergsten, C. Feasibility study for the implementation of an automatic system for the detection of social interactions in the waiting area of automatic milking stations by using a video surveillance system. Comput. Electron. Agric. 2016, 127, 506–509. [Google Scholar] [CrossRef]
- Guzhva, O.; Ardö, H.; Nilsson, M.; Herlin, A.; Tufvesson, L. Now You See Me: Convolutional Neural Network Based Tracker for Dairy Cows. Front. Robot. AI 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Jensen, D.B.; van der Voort, M.; Hogeveen, H. Dynamic forecasting of individual cow milk yield in automatic milking systems. J. Dairy Sci. 2018, 101, 10428–10439. [Google Scholar] [CrossRef]
- Jensen, D.B.; Hogeveen, H.; De Vries, A. Bayesian integration of sensor information and a multivariate dynamic linear model for prediction of dairy cow mastitis. J. Dairy Sci. 2016, 99, 7344–7361. [Google Scholar] [CrossRef]
- Ghiasi, H.; Piwczyński, D.; Khaldari, M.; Kolenda, M. ; And Application of classification trees in determining the impact of phenotypic factors on conception to first service in Holstein cattle. Anim. Prod. Sci. 2016, 56, 1061. [Google Scholar] [CrossRef]
- Piwczyński, D.; Sitkowska, B.; Kolenda, M.; Brzozowski, M.; Aerts, J.; Schork, P.M. Forecasting the milk yield of cows on farms equipped with automatic milking system with the use of decision trees. Anim. Sci. J. 2020, 91, e13414. [Google Scholar] [CrossRef]
- Kliś, P.; Sawa, A.; Piwczyński, D.; Sitkowska, B.; Bogucki, M. Prediction of cow's fertility based on data recorded by automatic milking system during the periparturient period. Reprod. Dom. Anim., 2021a, 56(9), 1227-1234. [CrossRef]
- Breiman, L., J.H. Friedman, R.A. Olshen, and C.J. Stone. Classification and Regression Trees. 1984. Wadsworth S, Pacific Grove, CA, USA.
- Kliś, P.; Piwczyński, D.; Sawa, A.; Sitkowska, B. Prediction of Lactational Milk Yield of Cows Based on Data Recorded by AMS during the Periparturient Period. Animals 2021, 11, 383. [Google Scholar] [CrossRef]
- Kadzere, C.; Murphy, M.; Silanikove, N.; Maltz, E. Heat stress in lactating dairy cows: a review. Livest. Prod. Sci. 2002, 77, 59–91. [Google Scholar] [CrossRef]
- Fuentes, S.; Viejo, C.G.; Cullen, B.; Tongson, E.; Chauhan, S.S.; Dunshea, F.R. Artificial Intelligence Applied to a Robotic Dairy Farm to Model Milk Productivity and Quality based on Cow Data and Daily Environmental Parameters. Sensors 2020, 20, 2975. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Banhazi, T.; Phillips, C.J.; Wang, C.; Li, B. A machine learning framework to predict the next month's daily milk yield, milk composition and milking frequency for cows in a robotic dairy farm. Biosyst. Eng. 2022, 216, 186–197. [Google Scholar] [CrossRef]
| Algorithm | Description |
|---|---|
| Regression analysis | Regression analysis is a statistical technique used to describe the relationships between variables. It allows predicting certain characteristics of output values based on input values [30]. It includes classical models such as simple and multiple linear regression, logistic regression, Generalized Linear Models (GLM), Generalized Additive Models (GAM), linear mixed models, polynomial regression, and time series. |
| Decision Tree (DT) |
A decision tree (DT) is a predictor that associates the features values with a label of a data instance by traveling from a root node to a leaf of a tree structure. Each node represents the splitting of the input space [29]. The feature or the value to be used for this splitting depends on the problem. A common splitting rule is the maximization of the Information Gain, which is the reduction in information entropy on splitted groups. When used for regression problems, it is called CART, an acronym for Classification and Regression Trees. |
| Random Forest (RF) | Random Forest (RF) is an ensemble classification model that combines several randomized decision trees. These decision trees are models that classify random subsets of the data where each subset contains responses of one class (either “yes” or “no”) [31]. Additionally, different trees can also use different sets of features to be trained or different random subsets of the data. For the RF outcome, the decision trees predictions are combined in a disambiguation method, as for example averaging [32] in the case of regression problems or major |
| AdaBoost | AdaBoost stands for Adaptive Boosting. It is also referred generically as Gradient Boosting. It combines sequentially the result of many weak decision trees. The first decision tree takes the raw data as input. The others receive as input the data weighted by the prediction errors of the previous classifier. Thus, each decision tree will adjust the prediction of the previous classifier. |
| k-Nearest Neighbors (k-NN) | The k-Nearest Neighbors (k-NN) algorithm looks at labeled points nearby an unlabeled point and based on this, to make a prediction of what the label should be [33]. Therefore, the learning strategy of k-NN is memorizing instead of finding relationships among features. |
| Support Vector Machine (SVM) | The objective of the Support Vector Machine (SVM) algorithm is to find the boundaries that maximize the distance of a multi-dimensional plane that separates the classes to be modeled. It uses the geometrical properties of the data to build these multi-dimensional boundaries between data points in the feature space belonging to different classes [34]. |
| Bayesian Networks (BN) | Bayesian networks (BN) are a type of probabilistic graphical model that uses Bayesian inference for probability computations. Bayesian networks aim to model conditional dependence, and therefore causation, by representing conditional dependence by edges in a directed graph [35]. The Naïve version assumes independence amongst the features, while the Tree-Augmented version also allows modeling the dependency amongst the features themselves. |
| Neural Networks (NN) | A Neural Network (NN) is a model that can be composed of three types of layers: an input layer, hidden layers, and an output layer. All layers are composed of nodes, which are sometimes called neurons. The outputs of each neuron are transformed by a nonlinear function and then are fed into the subsequent layers, with different weights along the connections between the neurons. The input layer takes in some numerical representation of the data. The output layer produces a prediction. The hidden layers perform transformations on the data which are usually nonlinear [36]. Various types of NN include Multilayer Perceptron (MLP), Back Propagation Neural Network (BPNN or NN for short, since this is the most used neural network), Probabilistic Neural Network (PNN), Recurrent Neural Network (RNN), Convolutional Neural Network (CNN). A CNN is a Deep Learning algorithm, which means that its network architecture usually needs a high number of hidden layers. The CNN takes in an input image, and, in these hidden layers, it assigns importance (learnable weights and biases) to various aspects/objects in the image. Then, this transformed image is used for the classification in the output layer. |
| Self-Organizing Maps (SOM) | Self-Organizing Maps (SOM) are a different type of Neural Network. They show only one layer with a predefined number of nodes. These nodes are linked to the input data and the value associated to these connections represent the distance between them. Thus, the SOM can be seen as a two-dimensional representation of the data, in which the data structure is preserved. The most common use of this algorithm is in clustering analysis, which requires a post-processing phase in which de SOM nodes will be clustered. |
| Clustering Algorithms | Clustering algorithms aim at dividing objects into groups (clusters) using measures of similarity and dissimilarity among the objects. The goal is to maximize the similarity among objects of the same cluster and, at the same time, maximize the dissimilarity among objects that belong to different clusters. Examples of these measures are one minus correlation or Euclidean distance. Some of the most frequently used clustering techniques include hierarchical clustering and k-means clustering. Hierarchical clustering (HC) creates a hierarchical tree-like structure of the data in which the length of the branches reflects the dissimilarity among the two clusters that it separates. The hierarchical tree is cutted at some point and the branches that are separated at this cut will define the clusters of the objects. The k-means clustering algorithm starts with random cluster centers (k), being the number of these clusters specified by the user [26]. The data points are assigned to the nearest cluster center. The center clusters are, then, redefined according to the new cluster configuration. This process is repeated iteratively until the cluster centers are no longer modified or until a maximum number of iterations. |
| Fuzzy logic | In Fuzzy Logic theory, the objects do not belong exclusively to one set (or class) or to another. Instead, they have a continuum of grades of membership to all classes, varying from 0 to 1 [37]. Fuzzy logic-based decision support systems usually follow three basic steps. First, the input values are fuzzified by the assignment of the membership functions. Second, a set of logic rules are applied to transform the input fuzzy values, generating the fuzzy output. Lastly, these outputs are defuzzified to generate the crisp system prediction. It is one of the methods developed to achieve the optimum solution situations where the input variables relate to the output variables by means of highly non-linear relationships. As a special case, the Adaptive Neuro Fuzzy Inference System (ANFIS) is a NN to map numerical inputs into an output through fuzzy-based rules. |
| Genetic Algorithms (GA) | Genetic Algorithms (GA) are search algorithms of the family of Evolutionary Algorithms based on the mechanics of natural selection and natural genetics [38]. The individuals are the possible solutions for the problem to be optimized. The set of these individuals that evolve together form the algorithm's population, and the fitness of the individuals is the criteria for a probabilistic selection of the solutions, in which the better the fitness, the higher the probability of that individual to be selected to the next generation. This type of stochastic search algorithm is often used in ML applications [39]. GAs are used in discrete spaces and find their applications where other gradient-based methods cannot be used. A GA is best suited to situations where information is a critical criterion for performance [24]. |
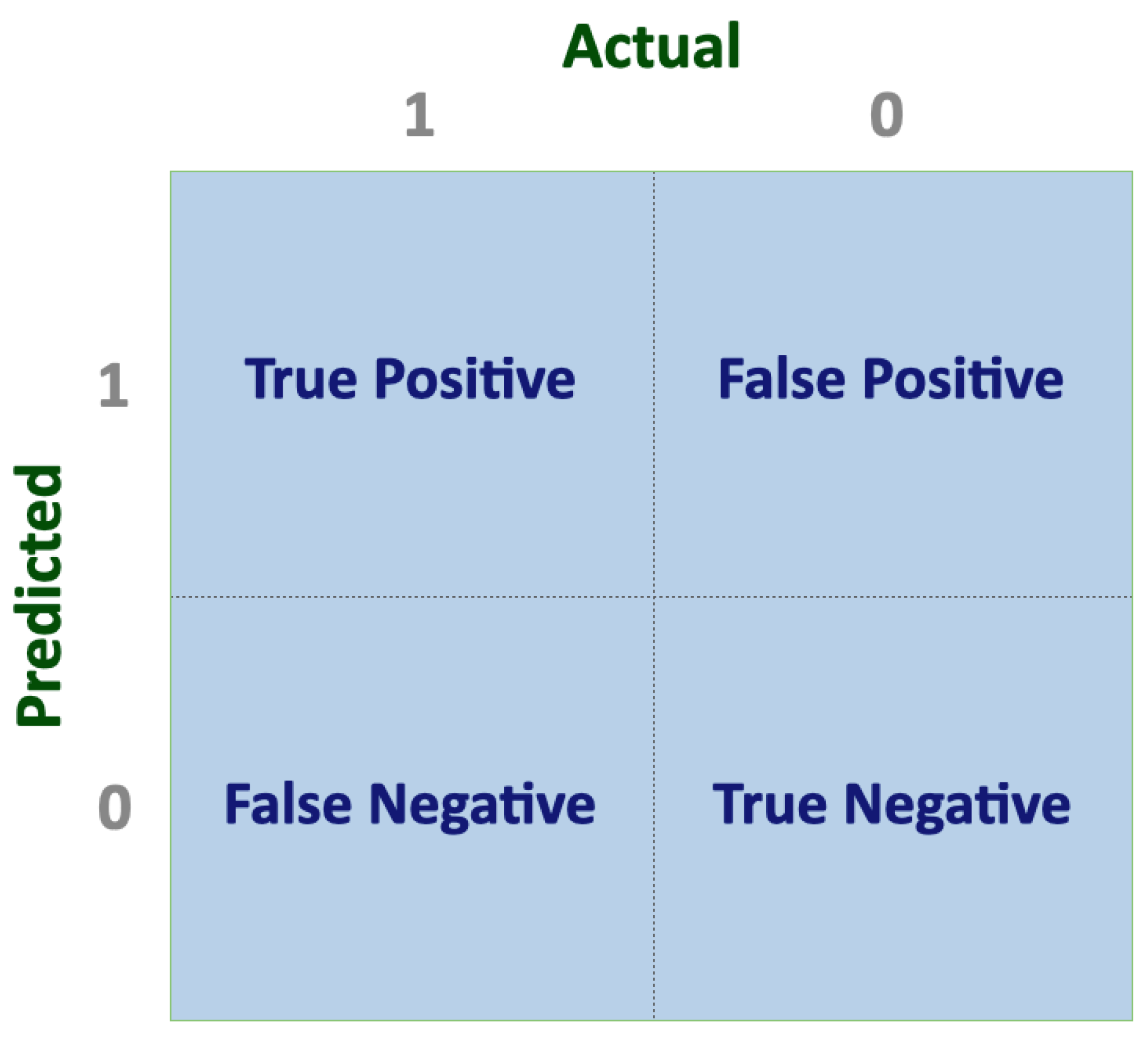
| Performance metric | Formula | Description |
|---|---|---|
| Accuracy | It is the ratio between the number of correct predictions versus the total number of inputs samples. | |
| Error rate | It is the ratio between the number of wrong predictions versus the total number of inputs samples. | |
| Sensitivity (Recall) | It measures the proportion of correctly identified positive examples. | |
| Specificity | It measures the proportion of correctly identified negative examples. | |
| Precision | It is the proportion of positive predictions that are correct. | |
| F1 score | It combines precision and sensitivity in a harmonic mean. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





