1. Introduction:
Tuberculosis is caused by
“Mycobacterium tuberculosis” which displays multidrug resistance to primary antibiotic treatment [
1]. The reasons for such resilient behavior are related to the unique physical, chemical, and biological makeup of this bacterium. It contains lipids that form a waxy impervious coat, preventing chemical interactions and even death [
2].
However, Trehalose Dimycolate, a glycolipid found on the cell surface, and its derivative analog trehalose dimycolate, have been shown to elicit an immunogenic response as they are bioactive and can react with mincle receptors on neighboring macrophages [
3,
4,
5]. In this project, we visually and statistically examined how exactly these glycolipids/ligands bind to the carbohydrate domain of the c-type lectin molecule using a process called molecular docking [
6,
7,
8,
9]. Docking is a computational procedure in which various software packages generate different positions at which ligands bind to their receptors [
9,
10,
11].
Molegro Virtual Studio and Discovery Studio Docking software were used in this study [
11]. ChemDraw and ChemDraw3D were used for ligand preparation and construction because protein preparation using docking software is time consuming. Trehalose dibehenate showed the highest affinity for binding to mincle and triggered the highest release of cytokines in terms of biological activity, as its poses had the most intense docking scores for various software programs [
1]. Images of these poses and interactions were captured and are presented in the results. Trehalose dibenzenate (TDB) can be used to manufacture drugs and as a vaccine adjuvant to trigger strong immune responses [
4].
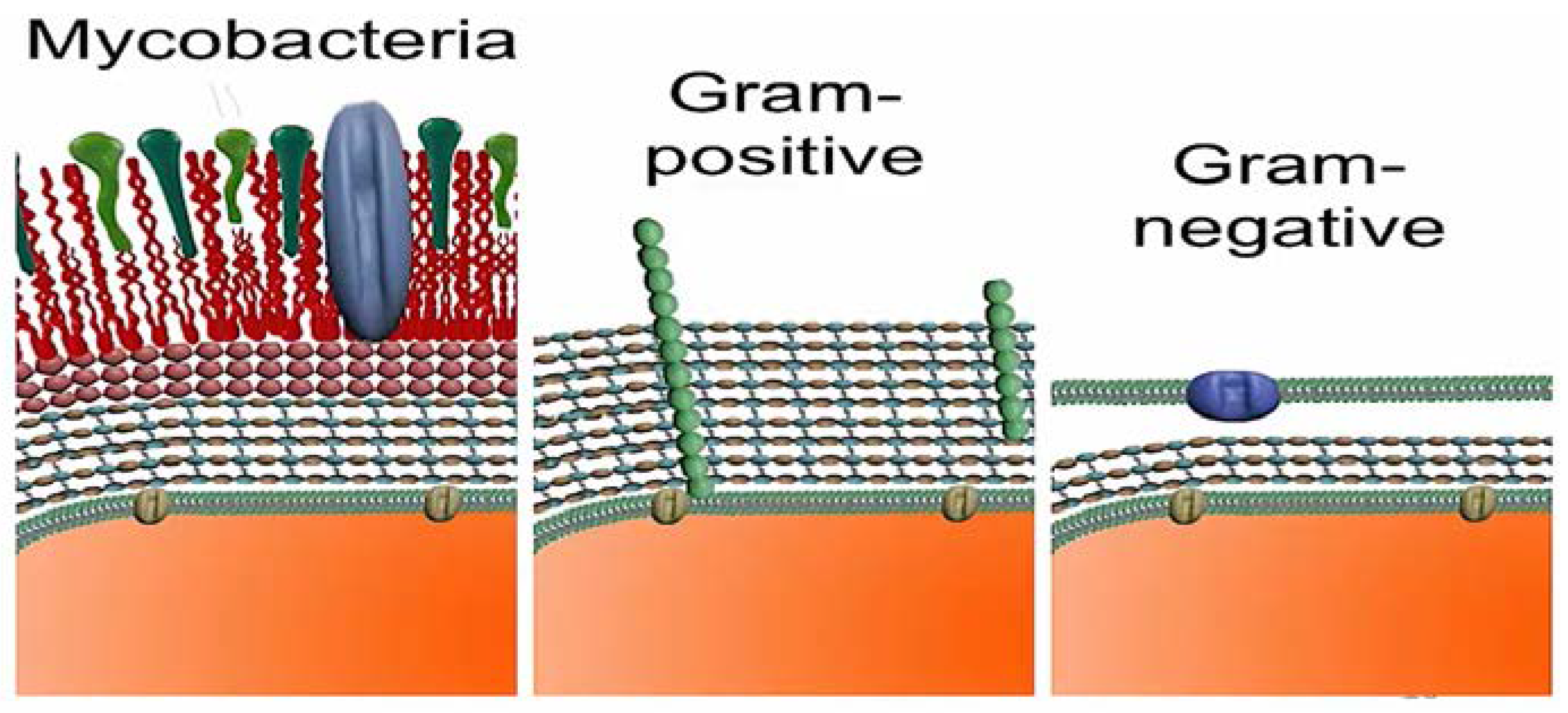
Mycobacteria (
Figure 1) lack an outer membrane resembling that of Gram-positive bacteria. However, as shown above, the peptidoglycan layer is not as thick as that of gram-positive bacteria [
1,
2].
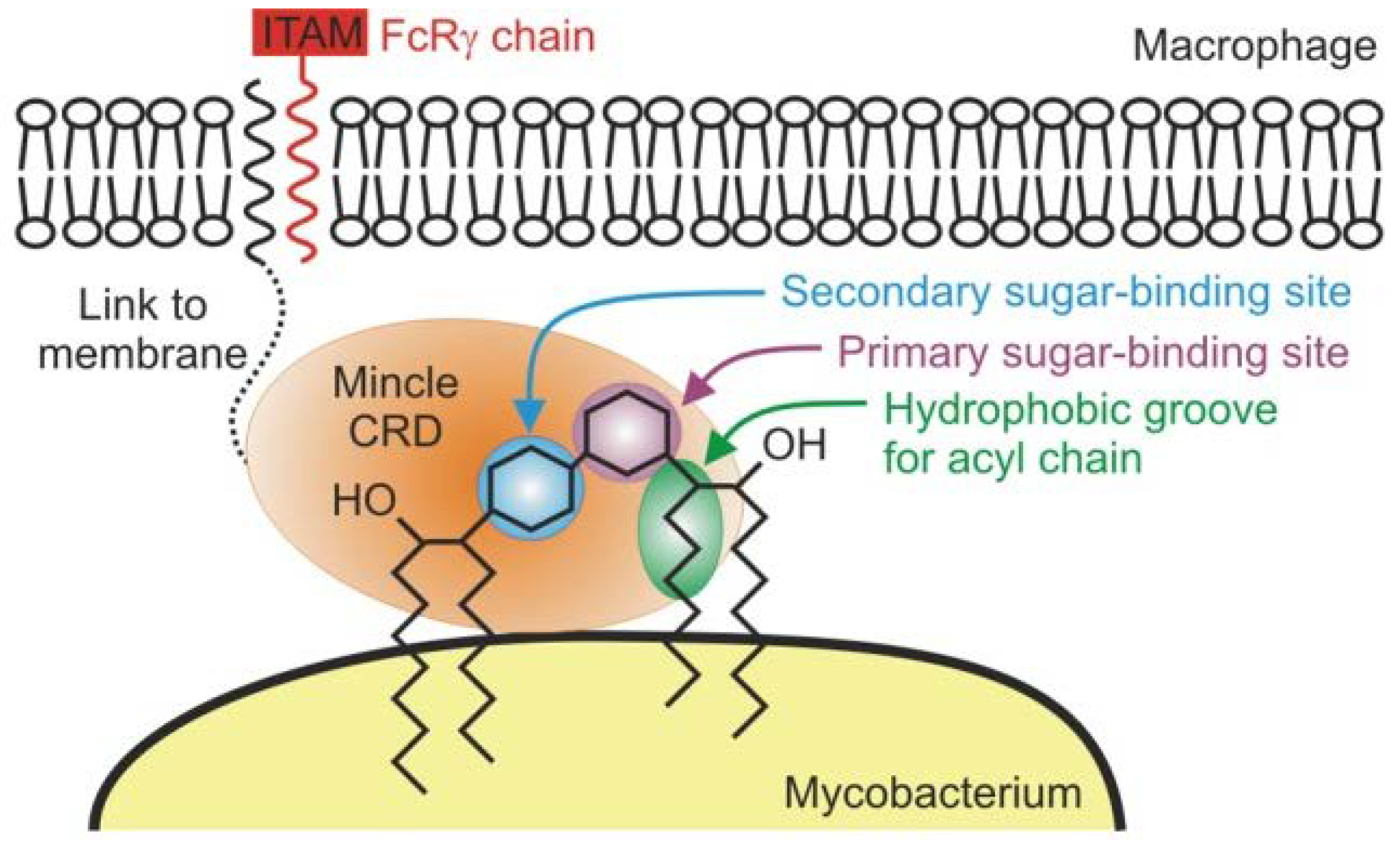
Mincle is a macrophage-inducible C-type lectin expressed on myeloid cell outskirts that recognizes damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) [
4]. The TDM molecule and its synthetic analog TDB act as ligands for mincle receptors and exhibit anticancer activity by activating the spleen tyrosine kinase (Syk) and Card9-Bc110-MALT1 inflammasome pathways [
12,
13]. Part of the receptor binds to the C-type carbohydrate domain, which is attached to the cell surface via a transmembrane anchor [
9,
13,
14,
15]. After binding, a hetero-oligomer is formed with the gamma subunit of the Fc receptor, which interacts with spleen tyrosine kinase (Syk) enzymes through the immunotyrosine activation motif (ITAM), leading to the CARD9 signaling pathway cascade. This, in turn releases TH1 and TH17 molecules that induce pro-inflammatory cytokines such as IL-1, IL-7, and TNF-α. This induces inflammation in the macrophages and dendritic cells. [
16]
The protein ligand interaction between (TDM) and (TDB) to its protein receptor molecule for the purposes of this research will be done using the modeling technique known as molecular docking (
Figure 2), which is used to predict how proteins interact with various molecules such as ligands. The aftermath of such interactions can result in a complex that enhances, activates, or inhibits biological functions [
9]. All docking software possess an algorithm to identify the active site, and it explores the best possible binding position of the ligands [
9,
11].
Some of the disadvantages of these techniques are:
2. Materials and Methods
Methodology
The protein data bank ID code for the carbohydrate domain of C-type lectin mincle was obtained [
3]. After downloading the protein receptor, the file was saved in the exact format (. pdb) using docking software. Several images of trehalose dibenzoate (TDB) and trehalose dimycolate (TDM) were obtained using Google Scholar and Google, respectively. Using ChemDraw Professional [
10], the shapes of the TDB and TDM ligands acquired online using the same software ensured that no errors were found in the structure. Subsequently, the structure is copied.
The copied structure was then pasted in CHEM3D to obtain the 3-Dimensional structure of the ligand. Following the 3-Dimensional conversion the MM2 function was selected, where the 3D shape was altered to minimize steric interaction and energy, showing the most feasible configuration of the ligand molecule.
Using the Molegro virtual docker [
11], the (4KZV) carbohydrate region domain of the protein receptor mincle was imported into the software. All water molecules and external hydrogen bonds were removed for a concise visualization of the binding site. One ligand (GLC-GLC) was already present in the receptor and removed, allowing only two ligands to be present: TDB and TDM. The TDB molecule was imported as the ligand. The preparation option was selected for small molecules as the ligand was prepared for docking. The authors selected the preparation and detected all the cavities. A docking and docking wizard was selected, which allowed it to choose the ligand to dock, allowing for five possible poses and running the docking simulation. It saved the energy values and rearranged them in order of the most negative docking score. The moldock energy values are tabulated for each pose. This procedure was repeated for TDM.
Procedure for Docking Using Discovery Studios
After obtaining the configured files of the ligand from CHEMDRAW in a “cdxml” file format, using sourceforge.net to download open babel in which was used to convert the file format into a pdb format to be used in discovery studios docker, the 4KZV protein file and TDB files were imported separately. A TDB file was prepared for the ligand and was used as the input ligand. The protein was prepared, and all water molecules were removed. Using the libdock function, docking was started with the prepared molecules, five out of the 11 poses were made visible for viewing. The energy values were calculated and libdock values were obtained. The docking view was then converted into a two-dimensional image to view the interactions with the ligand [
9]. Subsequently, the values and images were recorded.
3. Results
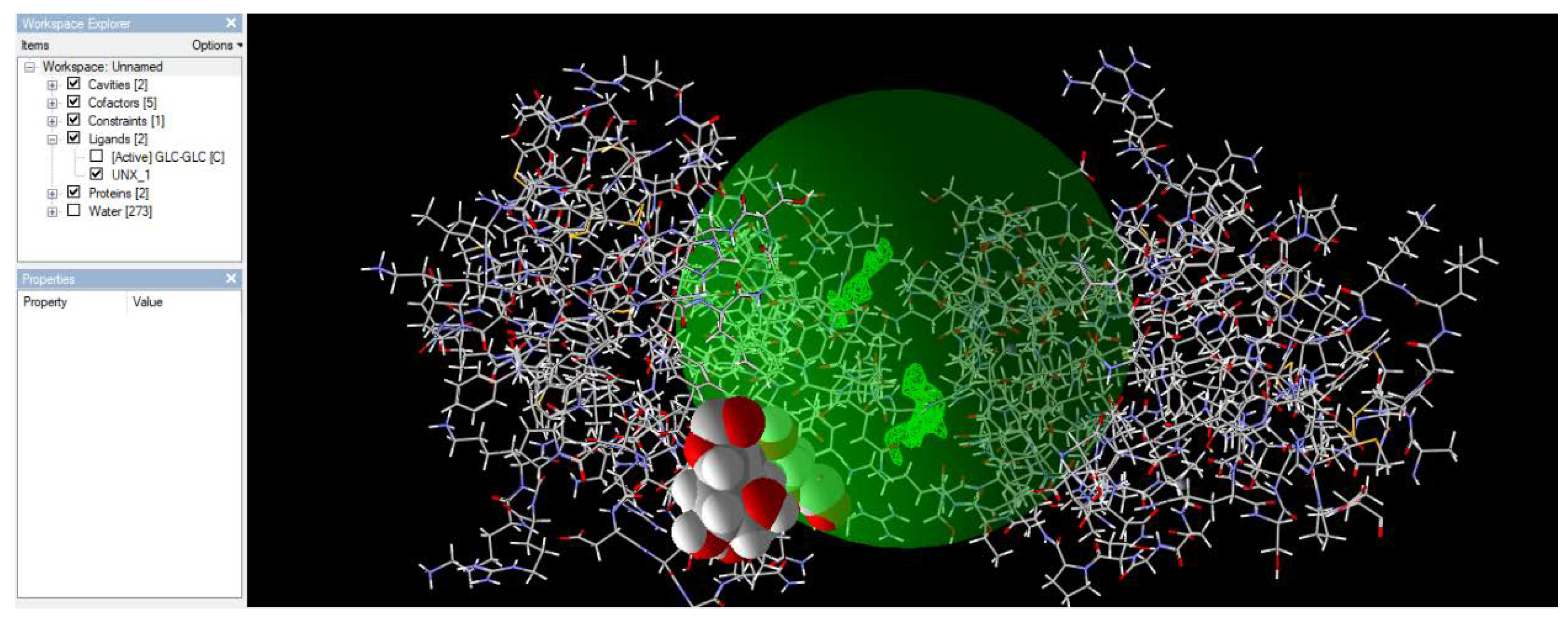
3.1.1. Docking of Mincle and TDM
Figure 3.
shows a feasible pose in which trehalose dimycolate (TDM) binds to the mincle receptor.
Figure 3.
shows a feasible pose in which trehalose dimycolate (TDM) binds to the mincle receptor.
Figure 3 shows a feasible pose in which trehalose dimycolate (TDM) binds to the mincle receptor. Among the five poses found, this pose had the lowest energy value, indicating minimal energy, as shown in nature.
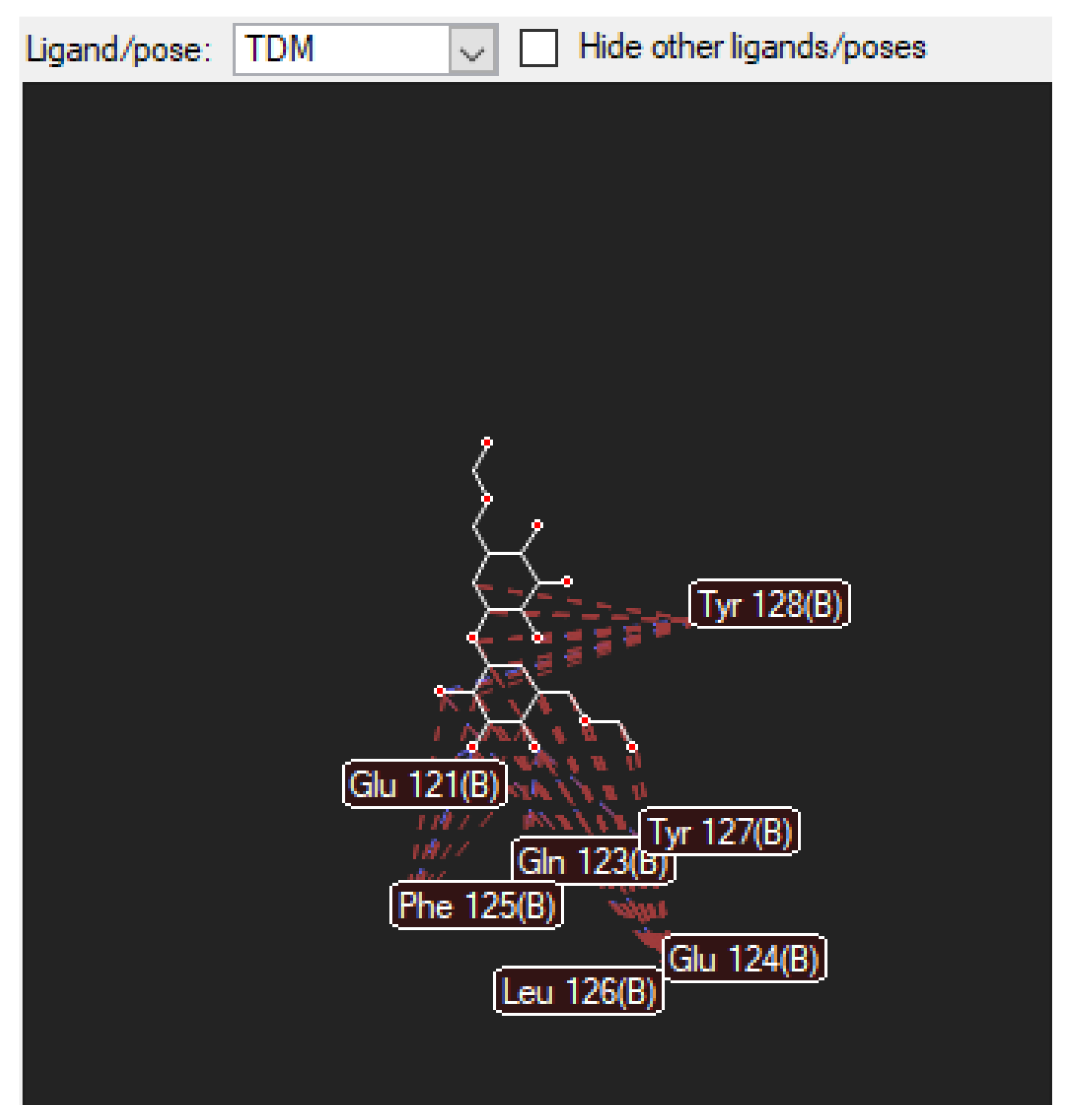
3.1.2. The Nearby Amino Acids in Which the TDM Binds to
Figure 4.
shows the diagram above illustrates the van der Waals and hydrogen bond-to-bond interactions between TDM and (as shown above) amino acids. These amino acids are found on the mincle receptor, which is the orientation in which they interact with the TDM ligand.
Figure 4.
shows the diagram above illustrates the van der Waals and hydrogen bond-to-bond interactions between TDM and (as shown above) amino acids. These amino acids are found on the mincle receptor, which is the orientation in which they interact with the TDM ligand.
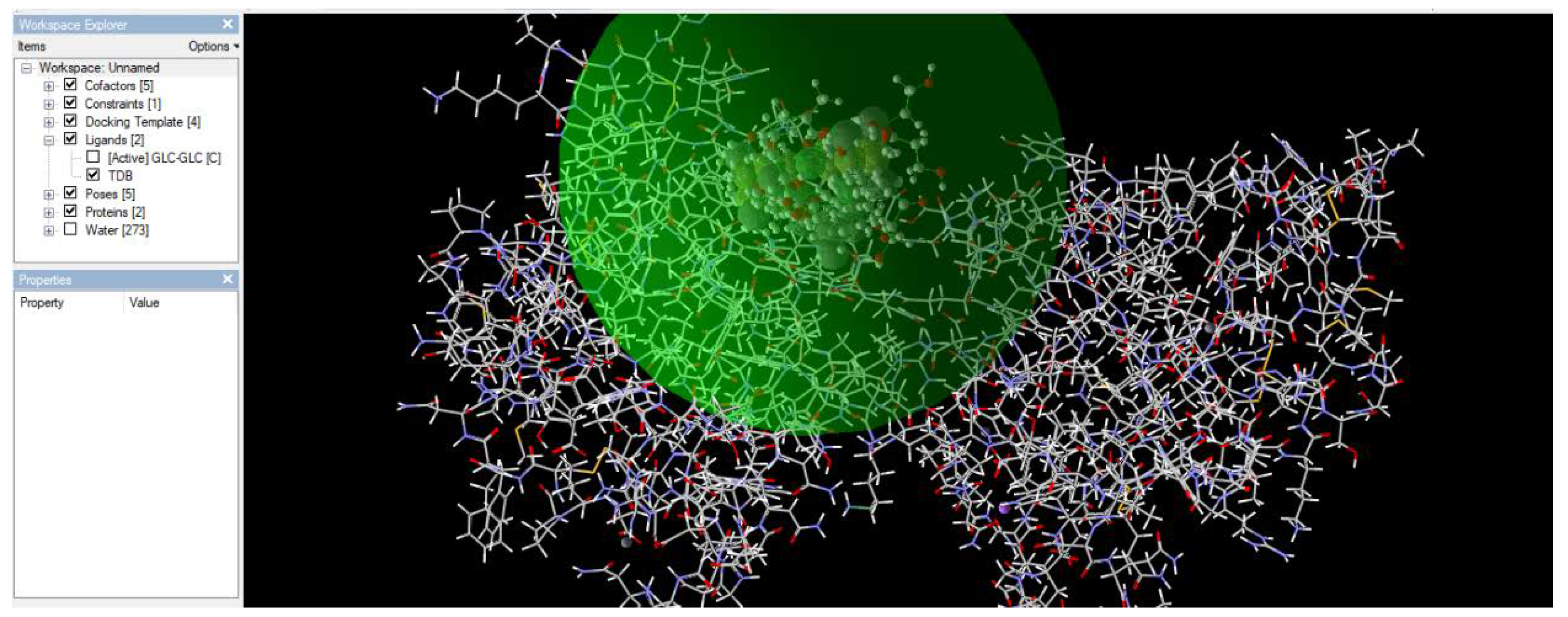
3.2.1. Docking of Mincle and TDB
Figure 5.
shows a feasible pose in which the TDB binds to the mincle receptor. Among the five poses found, this pose has the lowest energy value, indicating minimal energy, as shown in nature.
Figure 5.
shows a feasible pose in which the TDB binds to the mincle receptor. Among the five poses found, this pose has the lowest energy value, indicating minimal energy, as shown in nature.
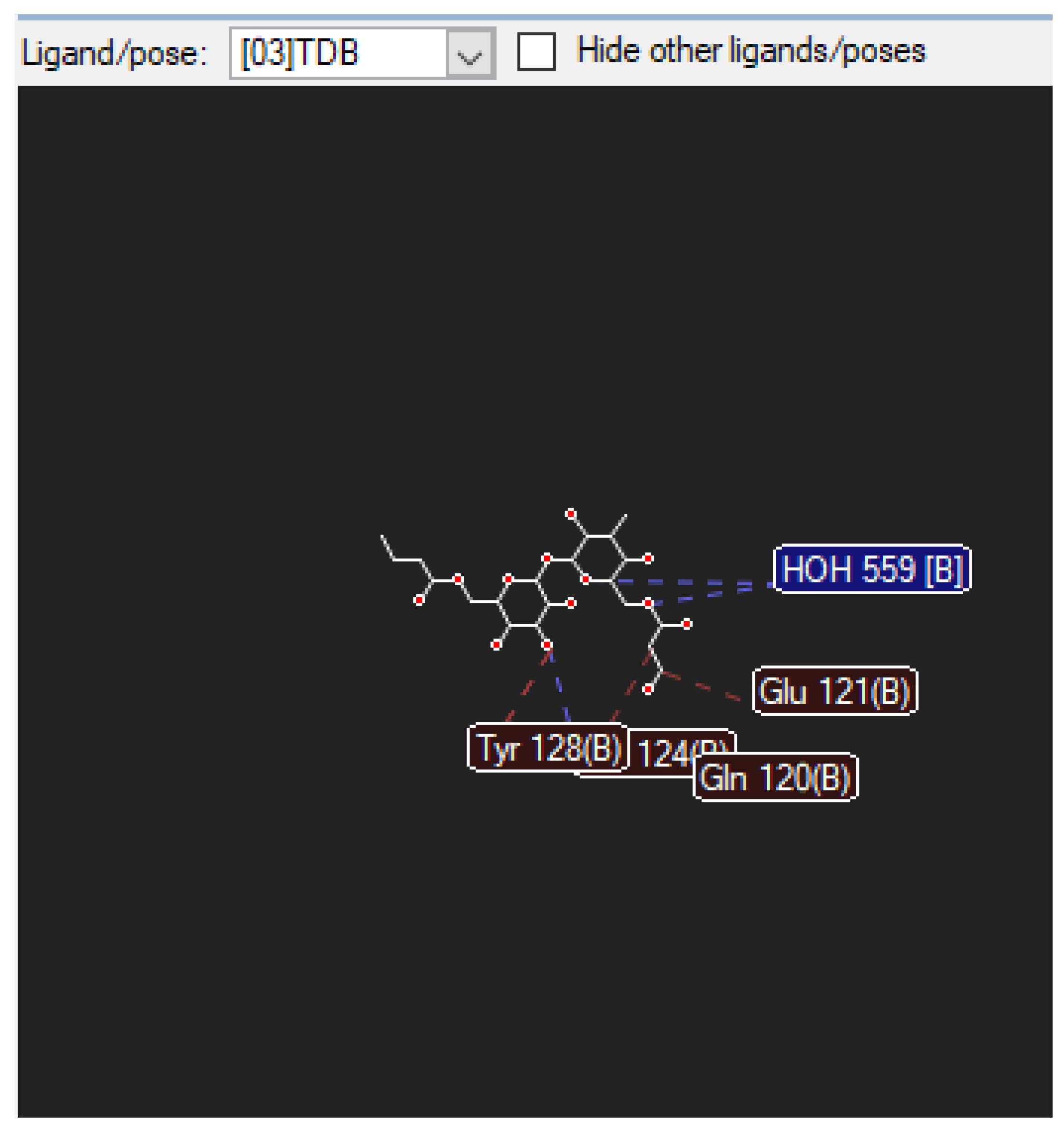
3.2.2. The Nearby Amino Acids Which TDM Binds to
Figure 6.
illustrates the van der Waals and hydrogen bond-to-bond interactions between TDB and (as shown above) amino acids. These amino acids are found on the mincle receptor, which is the orientation in which they interact with the TDB ligand.
Figure 6.
illustrates the van der Waals and hydrogen bond-to-bond interactions between TDB and (as shown above) amino acids. These amino acids are found on the mincle receptor, which is the orientation in which they interact with the TDB ligand.
Table 1.
Showing energy values using the molegro studios virtual docker for TDB.
Table 1.
Showing energy values using the molegro studios virtual docker for TDB.
| POSE |
LIGAND |
MolDock SCORE |
Rerank Score |
Docking score |
| 4 |
TDB |
-40.4109 |
-31.3595 |
-402.02 |
| 3 |
TDB |
-36.6981 |
2.50891 |
-410.682 |
| 2 |
TDB |
-24.9085 |
-9.17407 |
-414.356 |
| 5 |
TDB |
-24.6662 |
-30.2681 |
-401.288 |
| 1 |
TDB |
-16.5354 |
61.5971 |
-442.523 |
Table 1 lists the results of the calculations performed using Molegro Studio virtual docker. This software provides a forcefield-generated energy value in the form of a “moledock score, where the more negative the a value, the more likely it is to exist in nature. Therefore, from the results, pose number 4 with a value of -40.4109 has the highest chance of existing biologically, whereas pose number 1 with a value of -16.5354 is the least likely to exist in nature, as it requires too much energy to do so.
Table 2.
Showing energy values using the molegro studios virtual docker for TDM.
Table 2.
Showing energy values using the molegro studios virtual docker for TDM.
| POSE |
LIGAND |
MolDock SCORE |
Rerank Score |
Docking score |
| 3 |
TDM |
-42.1683 |
-38.2711 |
-261.883 |
| 4 |
TDM |
-34.7130 |
-16.1923 |
-247.830 |
| 1 |
TDM |
-30.0261 |
-4.84853 |
-268.438 |
| 2 |
TDM |
-27.6750 |
-17.0551 |
-265.637 |
| 5 |
TDM |
-2.75511 |
-28.6257 |
-244.610 |
Table 2 shows the results of the calculations performed using Molegro Studio virtual docker. This software provides a forcefield-generated energy value in the form of a “moledock score, where the more negative the a value, the more likely it is to exist in nature. Therefore, from the results, pose number 3 with a value of -42.1683 holds the highest chance of existing biologically, whereas pose number 5 with a value of -2.75511 is the least likely pose to exist in nature, as it requires too much energy to do so.
Table 3.
Showing docking scores using Discovery studios docker.
Table 3.
Showing docking scores using Discovery studios docker.
| POSE No. |
LIGAND |
LibDOCK SCORE |
| 6 |
TDB |
89.9593 |
| 5 |
TDB |
96.6506 |
| 4 |
TDB |
99.7128 |
| 3 |
TDB |
116.655 |
| 1 |
TDB |
124.043 |
For this particular part of the research experiment, discovery studios, docker is used and its docking score or energy value for each individual pose is converted to a “libdock score” where a higher reading would signify a greater energy requirement for natural existence (table 3). Therefore, we can say that pose 6 values of 89.9593 would require the least amount of energy to exist in such a pose. However, the pose number 1 value at 124.043 would require the highest amount of energy to exist in nature, and is therefore almost enviable.
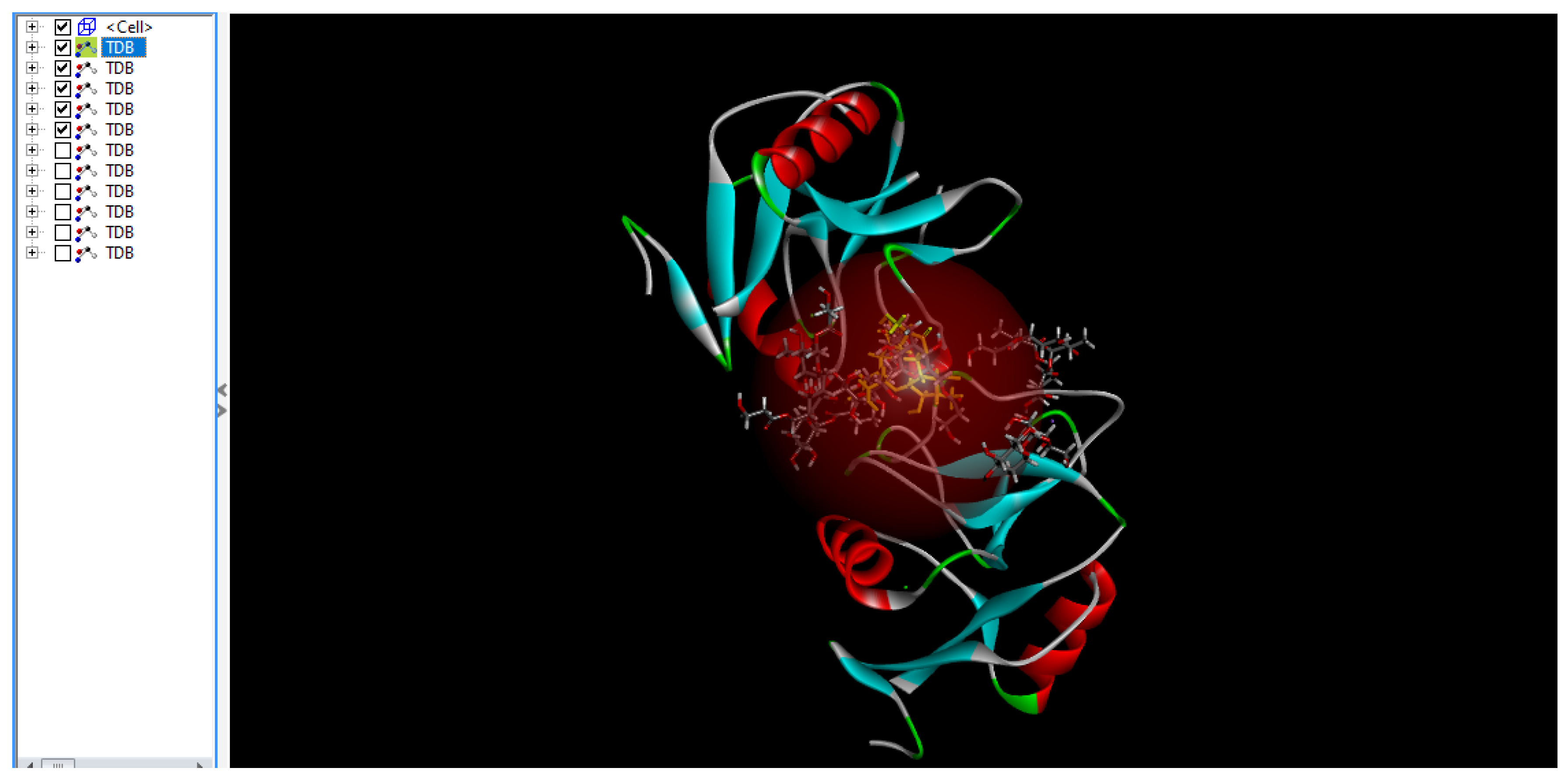
Figure 7.
Diagram above illustrates all 5 poses are connected.
Figure 7.
Diagram above illustrates all 5 poses are connected.
The diagram above (
Figure 7) shows the five most feasible poses in which TDB, the derivative analog of TDM, binds to mincle because of the low force field energy shown above in a reddish hemisphere, which is the area in which most hydrogen bond interactions occur.
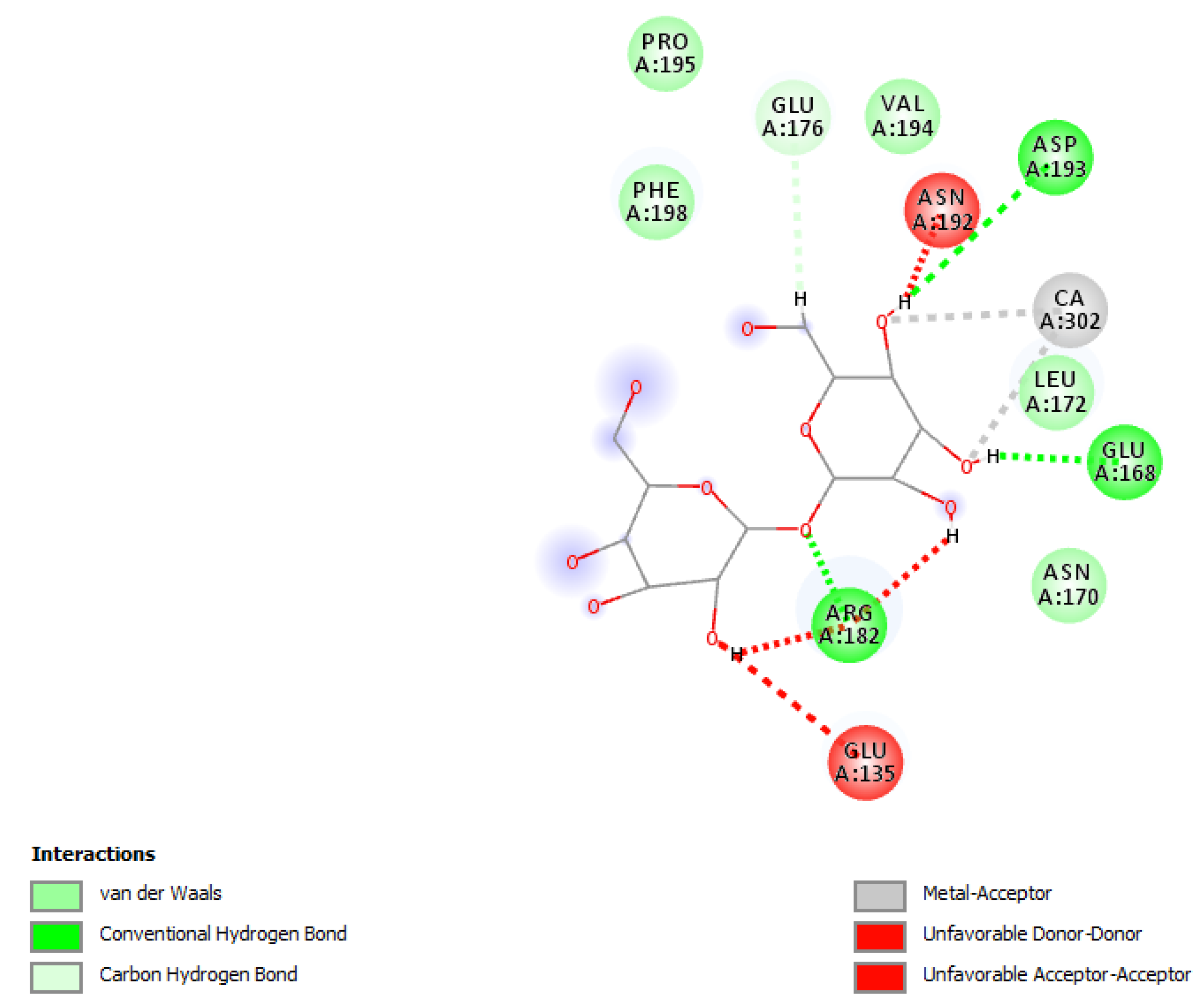
Figure 8.
The most feasible ligand poses interacting with molecules to bind.
Figure 8.
The most feasible ligand poses interacting with molecules to bind.
For this diagram (
Figure 8), all five poses presented in the previous diagram illustrate that this pose is only feasible or energy viable because of amino acid interactions and hydrogen bonding with the TDB molecule. The scale underneath shows the different types of bonding, not just hydrogen bonding, but also van der Waals, conventional hydrogen bonds, carbon hydrogen bonds, metal acceptors, unfavorable donor donors, and unfavorable acceptors.
4. Discussion
In this study, molecular docking of tuberculosis proteins was conducted using Molegro virtual docker and Discovery Studio docking. Docking is a computational study used to determine the position or manner in which TDB and TDM bind to the carbohydrate domain of mincle. These values explain why POSE 1 of TDB has a higher affinity for binding to mincle and releasing cytokines than that of TDM in both Docking Studies and Biological activity studies [
6,
9]. For this research, I chose to draw my ligands using ChemDraw and optimized them using ChemDraw3D, despite it being a longer and tedious route. However, this has enabled me to view and examine my molecular interactions. The use of discovery studios for docking is a lengthy and time-consuming process for the preparation of protein receptors. Obtaining docking scores and biological activity data from online sources is difficult, because this is a specific and targeted study. Therefore, results showing the release of inflammatory cytokines directly related to the binding and activation of the mincle receptor were used [
18].
The Docking score used for the Molegro Virtual Studios Docker was a generated value for the binding energies, which is a determinant of two factors.
Force field based
Empirical scoring functions [
9,
19].
A lower or higher negative docking score indicates less energy in the pose [
11,
16]. The other criterion was used to predict affinity. This score includes hydrogen bonding and electrostatic terms, which the scoring function does not, and a more positive value indicates better outcome. Furthermore, there are other criteria and yet another way to generate accurate results: Molegro’s re-ranking score, which combines both and, produces a value that signifies the best possible docking pose. According to the data in
Table 1 and
Table 2, pose 1 had the highest values of 69.5971 and -4.84853 respectively as their docking diagrams and interactions are shown [
11,
20].
For Discovery Studios, the docking and calculations in this study were performed with LibDock, which combines the values for hydrogen bonding, van der Waals forces of attraction, pi interactions, and other values to generate a score that indicates a high probability of ligand-receptor binding as the LibDock score increases [
21]. As shown in
Table 3, Pose 1 had the highest score of 124.03 as its docking diagram is highlighted in yellow, followed by a diagram showing its interactions. [
9].
In biological assays, TDM and TDB are detected by macrophages in the body, which ultimately leads to CARD9 signaling, thereby producing pro-inflammatory cytokines and chemokines. Therefore, by determining the highest concentrations of cytokine production, one can determine which ligand and concentration results in the highest activation and can be used therapeutically [
1,
9,
21].
5. Conclusions
In retrospect, it was possible to perform molecular docking of carbohydrates with M. tuberculosis proteins. The newly identified role of the Mincle and Syk/Card9-coupled signaling axis in infections could provide novel targets for the treatment of patients with tuberculosis. One disadvantage of Molegro studio docking is that it does not show the metallic atoms with which it interacts but rather shows hydrogen bonding and amino acid interactions.
Author Contributions
Conceptualization, S.S. and N.J.; methodology and software; validation, S.S., N.J., and A.J.V; formal analysis, S.S and N.J.; investigation, S.S., N.J., and A.J.V.; resources, S.S. and A.J.V; writing—original draft preparation, S.S. and A.J.V; writing—review and editing, S.S. and A.J.V; N.J.; supervision, N.J. and A.J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This study did not receive any external funding
Institutional Review Board Statement
The study did not require ethical approval.
Data Availability Statement
“Not applicable”.
Acknowledgments
To the West Indian Immunology Society for source editing.
Conflicts of Interest
“The authors declare no conflict of interest.”.
References
- Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG. MARCO, TLR2, and CD14 are required for macrophage cytokine response to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog. 2009 Jun;5(6):e1000474. [CrossRef]
- Correia-Neves M, Nigou J, Mousavian Z, Sundling C, Källenius G. Immunological hyporesponsiveness in tuberculosis: The role of mycobacterial glycolipids. Front Immunol. 2022 Dec 2;13:1035122. [CrossRef]
- Dohnálek J, Skálová T. C-type lectin-(like) fold-protein-protein interaction patterns and utilization. Biotechnol Adv. 2022 Sep;58:107944. [CrossRef]
- Ostrop J, Jozefowski K, Zimmermann S, et al.: Contribution of MINCLE-SYK signaling to activation of primary human APCs by mycobacterial cord factor and the novel adjuvant TDB. J Immunol. 2015 Sep 1;195(5):2417-28. [CrossRef]
- Kod Huber A, Kallerup RS, Korsholm KS, Franzyk H, Lepenies B, Christensen D, Foged C, Lang R. Trehalose diester glyolipids are superior to the monoesters in binding to Mincle, activation of macrophages in vitro and adjuvant activity in vivo. Innate Immun. 2016 Aug;22(6):405-18. Epub 2016 Jun 1. PMID: 27252171; PMCID: PMC5098696. [CrossRef]
- Allaka TR, Katari NK, Veeramreddy V et al.. Molecular Modeling of novel fluoroquinolone molecules. Curr Drug Discov Technol. 2018;15(2):109-122. [CrossRef]
- Foster AJ, Nagata M, Lu X, Lynch AT, Omahdi Z, Ishikawa E, Yamasaki S, Timmer MSM, Stocker BL. Lipidated Brartemicin Analogs are Potent Th1-Stimulating Vaccine Adjuvants. J Med Chem. 2018 Feb 8;61(3):1045-1060. [CrossRef]
- Gupta CL, Babu Khan M, Ampasala DR, Akhtar S, Dwivedi UN, Bajpai P. Pharmacophore-based virtual screening approach for identification of potent natural modulatory compounds of human Toll-like receptor 7. J Biomol Struct Dyn. 2019 Nov;37(18):4721-4736. [CrossRef]
- Saikia S, Bordoloi M. Molecular Docking: Challenges, Advances and its Use in Drug Discovery Perspective. Curr Drug Targets. 2019;20(5):501-521. [CrossRef]
- Pilipović A, Mitrović D, Obradović S, Poša M. Docking-based analysis and modeling of the activity of bile acids and their synthetic analogues on large conductance Ca2+ activated K channels in smooth muscle cells. Eur Rev Med Pharmacol Sci. 2021 Dec;25(23):7501-7507. [CrossRef]
- Mathpal D, Masand M, Thomas A, Ahmad I, Saeed M, Zaman GS, Kamal M, Jawaid T, Sharma PK, Gupta MM, Kumar S, Srivastava SP, Balaramnavar VM. Pharmacophore modeling, docking and the integrated use of a ligand- and structure-based virtual screening approach for novel DNA gyrase inhibitors: synthetic and biological evaluation studies. RSC Adv. 2021 Oct 25;11(55):34462-34478. [CrossRef]
- Pedersen GK, Andersen P, Christensen D. Immunocorrelates of CAF family adjuvants. Semin Immunol. 2018 Oct;39:4-13. Epub 2018 Nov 2. PMID: 30396811. [CrossRef]
- Zhao XQ, Zhu LL, Chang Q, Jiang C, You Y, Luo T, Jia XM, Lin X. C-type lectin receptor dectin-3 mediates trehalose 6,6’-dimycolate (TDM)-induced Mincle expression through CARD9/Bcl10/MALT1-dependent nuclear factor (NF)-κB activation. J Biol Chem. 2014 Oct 24;289(43):30052-62. [CrossRef]
- Miyake Y, Yamasaki S. Immune Recognition of Pathogen-Derived Glycolipids Through Mincle. Adv Exp Med Biol. 2020;1204:31-56. [CrossRef]
- Zhang D, Jia Y, Sun X, Li H, Yin M, Li H, Dai L, Han L, Wang L, Qian M, Du J, Zhu K, Bao H. The Dectin-1 Receptor Signaling Pathway Mediates the Remyelination Effect of Lentinan through Suppression of Neuroinflammation and Conversion of Microglia. J Immunol Res. 2022 Dec 28;2022:3002304. [CrossRef]
- Tam JM, Reedy JL, Lukason DP, Kuna SG, Acharya M, Khan NS, Negoro PE, Xu S, Ward RA, Feldman MB, Dutko RA, Jeffery JB, Sokolovska A, Wivagg CN, Lassen KG, Le Naour F, Matzaraki V, Garner EC, Xavier RJ, Kumar V, van de Veerdonk FL, Netea MG, Miranti CK, Mansour MK, Vyas JM. Tetraspanin CD82 Organizes Dectin-1 into Signaling domains to mediate cellular responses to Candida albicans. J Immunol. 2019 Jun 1;202(11):3256-3266. [CrossRef]
- Schwarz H, Posselt G, Wurm P, Ulbing M, Duschl A, Horejs-Hoeck J. TLR8 and NOD signaling synergistically induce the production of IL-1β and IL-23 in monocyte-derived DCs and enhance the expression of the feedback inhibitor SOCS2. Immunobiology. 2013 Apr;218(4):533-42. [CrossRef]
- Antas PRZ, da Silva ASM, Albuquerque LHP, Almeida MR, Pereira ENGS, Castello-Branco LRR, de Ponte CGG. The BCG Moreau Vaccine Upregulates In Vitro the Expression of TLR4, B7-1, Dectin-1 and EP2 on Human Monocytes. Vaccines (Basel). 2022 Dec 30;11(1):86. [CrossRef]
- Feinberg H, Jégouzo SAF, Rex MJ, Drickamer K, Weis WI, Taylor ME. Mechanism of pathogen recognition by human dectin-2. J Biol Chem. 2017 Aug 11;292(32):13402-13414. [CrossRef]
- Alam S, Khan F. Virtual screening, Docking, ADMET and System Pharmacology studies on Garcinia caged Xanthone derivatives for Anticancer activity. Sci Rep. 2018;8(1):5524. Published 2018 Apr 3. [CrossRef]
- Lee EJ, Brown BR, Vance EE, Snow PE, Silver PB, Heinrichs D, Lin X, Iwakura Y, Wells CA, Caspi RR, Rosenzweig HL. Mincle activation and the Syk/Card9 signaling axis are central to the development of autoimmune eye diseases. J Immunol. 2016 Apr 1;196(7):3148-58. Epub 2016 Feb 26. PMID: 26921309; PMCID: PMC4799727. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).