Submitted:
18 May 2023
Posted:
19 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Search Strategy and Selection Criteria
2.2. Data Extraction and Study Quality Assessment
2.3. Statistical Analysis
3. Results
3.1. Characteristics of the Included Studies
3.2. Decreased Alpha Diversity in ASD
3.3. Microbes Upregulated in ASD
3.4. Microbes Downregulated in ASD
3.5. Microbes with No Difference between ASD and TD
4. Discussion
Author Contributions
Acknowledgement
Conflicts of Interest
References
- American Psychiatric Association. The Diagnostic and statistical manual of mental disorders, 5th ed.; American Psychiatric Publishing Inc: Washington, DC, USA, 2013. [Google Scholar]
- Salari, N.; Rasoulpoor, S.; Rasoulpoor, S.; Shohaimi, S.; Jafarpour, S.; Abdoli, N.; Khaledi-Paveh, B.; Mohammadi, M. The global prevalence of autism spectrum disorder: A comprehensive systematic review and meta-analysis. Ital J Pediatr 2022, 48, 112. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Data & Statistics on Autism Spectrum Disorder 2022. Available online: https://www.cdc.gov/ncbddd/autism/data.html.
- Baron-Cohen, S.; Scott, F.J.; Allison, C.; Williams, J.; Bolton, P.; Matthews, F.E.; Brayne, C. Prevalence of autism-spectrum conditions: UK school-based population study. Br J Psychiatry 2009, 194, 500–509. [Google Scholar] [CrossRef] [PubMed]
- BLL, Y.S.K.; Koh, Y.-J.; Fombonne, E.; Laska, E.; Lim, E.-C.; Cheon, K.-A.; Kim, S.-J.; Kim, Y.-K.; Lee, H.; Song, D.-H.; Grinker, R.R. Prevalence of autism spectrum disorders in a total population sample. Am J Psychiatry 2011, 168, 904–912. [Google Scholar]
- Saito, M.; Hirota, T.; Sakamoto, Y.; Adachi, M.; Takahashi, M.; Osato-Kaneda, A.; Kim, Y.S.; Leventhal, B.; Shui, A.; Kato, S.; Nakamura, K. Prevalence and cumulative incidence of autism spectrum disorders and the patterns of co-occurring neurodevelopmental disorders in a total population sample of 5-year-old children. Mol Autism. 2020, 11, 35. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, X.; Yan, W.; Zou, X.; Wu, L.; Luo, X.; Li, T.; Huang, Y.; Guan, H.; Chen, X.; et al. Prevalence of Autism Spectrum Disorder in China: A Nationwide Multi-center Population-based Study Among Children Aged 6 to 12 Years. Neurosci Bull. 2020, 36, 961–971. [Google Scholar] [CrossRef]
- Holingue, C.; Newill, C.; Lee, L.C.; Pasricha, P.J.; Daniele Fallin, M. Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 2018, 11, 24–36. [Google Scholar] [CrossRef]
- Chaidez, V.; Hansen, R.L.; Hertz-Picciotto, I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord. 2014, 44, 1117–1127. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; Patterson, P.H.; Mazmanian, S.K. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Nadeem, A.; Ahmad, S.F.; Attia, S.M.; Al-Ayadhi, L.Y.; Al-Harbi, N.O.; Bakheet, S.A. Dysregulated enzymatic antioxidant network in peripheral neutrophils and monocytes in children with autism. Prog Neuropsychopharmacol Biol Psychiatry 2019, 88, 352–359. [Google Scholar] [CrossRef]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; Zheng, P. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci Rep. 2019, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Rattaz, C.; Michelon, C.; Munir, K.; Baghdadli, A. Challenging behaviours at early adulthood in autism spectrum disorders: Topography, risk factors and evolution. J Intellect Disabil Res. 2018, 62, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 2011, 108, 16050–16055. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Zegarra-Ruiz, D.F.; Kim, D.V.; Norwood, K.; Kim, M.; Wu, W.H.; Saldana-Morales, F.B.; Hill, A.A.; Majumdar, S.; Orozco, S.; Bell, R.; Round, J.L.; Longman, R.S.; Egawa, T.; Bettini, M.L.; Diehl, G.E. Thymic development of gut-microbiota-specific T cells. Nature 2021, 594, 413–417. [Google Scholar] [CrossRef]
- Ait-Belgnaoui, A.; Colom, A.; Braniste, V.; Ramalho, L.; Marrot, A.; Cartier, C.; Houdeau, E.; Theodorou, V.; Tompkins, T. Probiotic gut effect prevents the chronic psychological stress-induced brain activity abnormality in mice. Neurogastroenterol Motil. 2014, 26, 510–520. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. The impact of gut microbiota on brain and behaviour: Implications for psychiatry. Curr Opin Clin Nutr Metab Care. 2015, 18, 552–558. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; De Filippo, C. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017, 5, 24. [Google Scholar] [CrossRef]
- Ma, B.; Liang, J.; Dai, M.; Wang, J.; Luo, J.; Zhang, Z.; Jing, J. Altered Gut Microbiota in Chinese Children With Autism Spectrum Disorders. Front Cell Infect Microbiol. 2019, 9, 40. [Google Scholar] [CrossRef]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef]
- Li, D.; Chen, Q.; Zhuang, Z.; Chen, H. Correction between gut microbiota and symptom severity in children with autism spectrum disorder (in chinese). Chin J Child Health Care. 2023, 31, 497–501. [Google Scholar]
- Wong, O.W.H.; Lam, A.M.W.; Or, B.P.N.; Mo, F.Y.M.; Shea, C.K.S.; Lai, K.Y.C.; Ma, S.L.; Hung, S.F.; Chan, S.; Kwong, T.N.Y.; Wong, S.; Leung, P.W.L. Disentangling the relationship of gut microbiota, functional gastrointestinal disorders and autism: A case-control study on prepubertal Chinese boys. Sci Rep. 2022, 12, 10659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, H.; Tang, W.; Hao, X. Difference in fecal intestinal flora instructure and short chian fatty acids between children with autism spectrum disorder and healthy children(in Chinese). Chin J Child Health Care. 2022, 30, 1390–1394. [Google Scholar]
- Chen, Z.; Shi, K.; Liu, X.; Dai, Y.; Liu, Y.; Zhang, L.; Du, X.; Zhu, T.; Yu, J.; Fang, S.; Li, F. Gut Microbial Profile Is Associated With the Severity of Social Impairment and IQ Performance in Children With Autism Spectrum Disorder. Front Psychiatry. 2021, 12, 789864. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Yuan, B. Relationship between intestinal flora and autism grade in children with autism (in Chinese). Chin J Microecol. 2021, 33, 560–563. [Google Scholar]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; Qin, L.; Gu, B.; Liu, K.; Su, C.; Zhang, F.; Xia, Y.; Hu, Z.; Liu, X. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes. 2020, 11, 1246–1267. [Google Scholar] [CrossRef]
- Ding, X.; Xu, Y.; Zhang, X.; Zhang, L.; Duan, G.; Song, C.; Li, Z.; Yang, Y.; Wang, Y.; Wang, X.; Zhu, C. Gut microbiota changes in patients with autism spectrum disorders. J Psychiatr Res. 2020, 129, 149–159. [Google Scholar] [CrossRef]
- Zeng, J.; Wei, S.; Wang, M. Association between intestinal flora and symptom severity in children with autism spectrum disorders (in Chinese). J. Psychiatry. 2020, 33, 62–65. [Google Scholar]
- Zou, R.; Xu, F.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zhao, H.; Zheng, H. Changes in the Gut Microbiota of Children with Autism Spectrum Disorder. Autism Res. 2020, 13, 1614–1625. [Google Scholar] [CrossRef]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome. 2017, 5, 10. [Google Scholar] [CrossRef]
- Kang, D.W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe. 2018, 49, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism--comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Son, J.S.; Zheng, L.J.; Rowehl, L.M.; Tian, X.; Zhang, Y.; Zhu, W.; Litcher-Kelly, L.; Gadow, K.D.; Gathungu, G.; Robertson, C.E. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS ONE 2015, 10, e0137725. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef]
- Gondalia, S.V.; Palombo, E.A.; Knowles, S.R.; Cox, S.B.; Meyer, D.; Austin, D.W. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012, 5, 419–427. [Google Scholar] [CrossRef]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; Comegna, M.; Buommino, E.; Castaldo, G.; Bravaccio, C.; Chiariotti, L.; Berni Canani, R.; Lembo, F. Gut Microbiota Features in Young Children With Autism Spectrum Disorders. Front Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Pulikkan, J.; Maji, A.; Dhakan, D.B.; Saxena, R.; Mohan, B.; Anto, M.M.; Agarwal, N.; Grace, T.; Sharma, V.K. Gut Microbial Dysbiosis in Indian Children with Autism Spectrum Disorders. Microb Ecol. 2018, 76, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Inoue, R.; Sakaue, Y.; Sawai, C.; Sawai, T.; Ozeki, M.; Romero-Pérez, G.A.; Tsukahara, T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem. 2016, 80, 2450–2458. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Gomez-Fernandez, A.; Chueca, N.; Torre-Aguilar, M.J.; Gil, A.; Perez-Navero, J.L.; Flores-Rojas, K.; Martin-Borreguero, P.; Solis-Urra, P.; Ruiz-Ojeda, F.J.; Garcia, F.; Gil-Campos, M. Autism Spectrum Disorder (ASD) with and without Mental Regression is Associated with Changes in the Fecal Microbiota. Nutrients 2019, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, W.; Zhang, J.; He, Y.; Wang, J. Analysis of gut microbiota profiles and microbe-disease associations in children with autism spectrum disorders in China. Sci Rep. 2018, 8, 13981. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Vázquez, L.; Van Ginkel Riba, G.; Arija, V.; Canals, J. Composition of Gut Microbiota in Children with Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 792. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, X.; Li, J.; Li, F. Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Front Psychiatry 2019, 10, 473. [Google Scholar] [CrossRef]
- Parracho, H.M.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005, 54 Pt 10, 987–991. [Google Scholar] [CrossRef]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Sutterella Species, IgA-degrading Bacteria in Ulcerative Colitis. Trends Microbiol. 2020, 28, 519–522. [Google Scholar] [CrossRef]
- Xue, L.; He, J.; Gao, N.; Lu, X.; Li, M.; Wu, X.; Liu, Z.; Jin, Y.; Liu, J.; Xu, J.; Geng, Y. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep. 2017, 7, 45176. [Google Scholar] [CrossRef]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; de Vos, W.M.; Cani, P.D. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Berry, S.E.; Valdes, A.M.; Nguyen, L.H.; Piccinno, G.; Drew, D.A.; Leeming, E.; Gibson, R.; Le Roy, C.; Khatib, H.A.; et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021, 27, 321–332. [Google Scholar] [CrossRef]
| Study | Country | ASD | TD | Bacteria Detected | ||||
|---|---|---|---|---|---|---|---|---|
| n | Age(y) | male: female | n | age(y) | male: female | |||
| Li et al. (2023)[23] | China | 107 | 3.34 ± 0.75 | 79: 28 | 30 | 3.61 ± 0.48 | 21: 9 | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Verrucomicrobia |
| Wong et al.(2022)[24] | China | 92 | 8.43 ± 1.54 | 92: 0 | 112 | 8.12 ± 1.99 | 112: 0 | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteriota, Proteobacteria, VerrucomicrobiaGenus: Bacteroides, Bifidobacterium, Bilophila, Blautia, Collinsella, Dorea, Fusobacterium, Parabacteroides, Phascolarctobacterium, Sutterella |
| Zhang et al.(2022)[25] | China | 25 | 6.72 ± 2.56 | 18: 7 | 24 | 7.68 ± 2.93 | 14: 10 | Phylum: Actinobacteria, FirmicutesGenus: Bacteroides, Bifidobacterium, Blautia, Clostridium, Streptococcus, Sutterella |
| Chen et al.2021[26] | China | 138 | 6.11 ± 2.00 | 117: 21 | 60 | 6.65 ± 2.22 | 27: 33 | Genus:Bacteroides, Collinsella, Coprococcus, Faecalibacterium,Prevotella,Megamonas, Sutterella |
| Cheng et al.(2021)[27] | China | 36 | 7.44 ± 1.53 | 21: 15 | 50 | 7.33 ± 1.49 | 27: 23 | Genus: Bacteroides, Bifidobacterium, Clostridium |
| Table 1. Summary characteristics of the studies included in the meta-analysis (Cont.) | ||||||||
| Study | Country | ASD | TD | Bacteria Detected | ||||
| n | age(y) | male: female | n | age(y) | male: female | |||
| Dan et al.(2020)[28] | China | 143 | 4.94 ± 0.16 | 130:13 | 143 | 5.19 ± 0.17 | 127:16 | Phylum: Bacteroidetes, Cyanobacteria, FusobacteriaGenus: Blautia, Fusobacterium, Parabacteroides, Paraprevotella, Prevotella |
| Ding et al.(2020)[29] | China | 77 | 38.50±11.70m | 59: 18 | 50 | 42.9±14.5m | 39: 11 | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, VerrucomicrobiaGenus: Bacteroides, Bifidobacterium, Blautia, Collinsella, Dorea, Faecalibacterium,Paraprevotella, Streptococcus |
| Zeng et al.(2020)[30] | China | 90 | 3.50 ± 1.20 | 70: 20 | 50 | 3.00 ± 1.50 | 39: 11 | Phylum: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Fusobacteria, Tenericutes, Verrucomicrobia |
| Zou et al.(2020)[31] | China | 48 | 5 (2–7) | 38: 10 | 48 | 4 | 24: 24 | Genus:Akkermansia, Bacteroides, Megamonas, Prevotella |
| Ma et al.(2019)[21] | China | 45 | 7.27 ± 1.07 | 39: 6 | 45 | 7.04 ± 1.19 | 39: 6 | Phylum: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Fusobacteria, Proteobacteria, Tenericutes, Verrucomicrobia |
| Table 1. Summary characteristics of the studies included in the meta-analysis. (Cont.) | ||||||||
| Study | Country | ASD | TD | Bacteria Detected | ||||
| n | age(y) | male: female | n | age(y) | male: female | |||
| Ma et al.(2019)[21] | China | 45 | 7.27 ± 1.07 | 39: 6 | 45 | 7.04 ± 1.19 | 39: 6 | Genus: Bacteroides, Bifidobacterium, Bilophila, Faecalibacterium, Fusobacterium, Lactobacillus, Megamonas, Parabacteroides |
| Plaza et al.(2019)[45] | Spain | 30 | 44.19 ± 1.60m | - | 57 | 51±2.59 m | - | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia |
| Coretti et al.(2018)[41] | Italy | 11 | 35 ± 5.7m | 9: 2 | 14 | 35 ± 8.40m | 8: 6 | Genus: Bacteroides, Bifidobacterium, Blautia, Coprococcus, Streptococcus |
| Kang et al.(2018)[33] | USA | 23 | 10.1 ± 4.1 | 22:1 | 21 | 8.4 ± 3.4 | 15:6 | Genus: Akkermansia, Bacteroides, Coprococcus, Faecalibacterium, Prevotella |
| Pulikkan et al(2018)[43] | Indian | 30 | 9.5 (3–16) | 28: 2 | 24 | 9.5 (3.5–16) | 15: 9 | Phylum: Bacteroidetes, Firmicutes |
| Zhang et al.(2018)[46] | China | 35 | 4.9±1.5 | 29: 6 | 6 | 4.6±1.1 | 5: 1 | Phylum: Bacteroidetes, Firmicutes |
| Kang et al.(2017)[32] | USA | 18 | 7-16 | - | 20 | 7-16 | - | Genus: Bacteroides, Bifidobacterium, Clostridium, Coprococcus, Parabacteroides, Phascolarctobacterium |
| Table 1. Summary characteristics of the studies included in the meta-analysis. (Cont.) | ||||||||
| Study | Country | ASD | TD | Bacteria Detected | ||||
| n | age(y) | male: female | n | age(y) | male: female | |||
| Strati et al.(2017)[20] | Italy | 40 | 10 (5–17) | 31: 9 | 40 | 7 (3.6–12) | 28: 12 | Genus:Akkermansia,Bifidobacterium, Blautia, Collinsella, Coprococcus, Dorea, Faecalibacterium, Lactobacillus, Paraprevotella, Prevotella, Streptococcus |
| Inoue et al.(2016)[44] | Japan | 6 | 3-5 | - | 6 | 3-5 | - | Genus:Akkermansia, Bacteroides,Bifidobacterium, Bilophila, Blautia, Clostridium, Collinsella, Coprococcus, Dorea, Faecalibacterium |
| Son et al.(2015)[36] | USA | 34 | 7–14 | - | 31 | 7–14 | - | Phylum: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Proteobacteria, Tenericutes, Verrucomicrobia |
| Angelis et al.(2013)[42] | Italy | 10 | 4-10 | - | 10 | 4-10 | - | Genus:Bacteroides,Bifidobacterium, Clostridium, Faecalibacterium, Parabacteroides |
| Kang et al.(2013)[34] | USA | 20 | 6.70 ± 2.70 | 18: 2 | 20 | 8.30 ± 4.40 | 17: 3 | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia |
| Wang et al.(2013)[38] | Austrilia | 23 | 10.25 ± 0.75 | 21: 2 | 9 | 9.50 ± 1.25 | 4: 5 | Phylum: Actinobacteria, Bacteroidetes, Firmicutes, ProteobacteriaGenus: Sutterella |
| Table 1. Summary characteristics of the studies included in the meta-analysis. (Cont.) | ||||||||
| Study | Country | ASD | TD | Bacteria Detected | ||||
| n | age(y) | male: female | n | age(y) | male: female | |||
| Gondalia et al(2012)[40] | Australia | 28 | 2-12 | - | 25 | 2-12 | - | Genus:Bacteroides,Bifidobacterium, Coprococcus, Faecalibacterium, Parabacteroides, Phascolarctobacterium |
| Adams et al.(2011)[35] | USA | 58 | 6.91 ± 3.40 | 50: 8 | 39 | 7.70 ± 4.40 | 18: 21 | Genus: Bifidobacterium, Lactobacillus |
| Wang et al.(2011)[39] | Austrilia | 23 | 10.25 ± 0.75 | 21: 2 | 9 | 9.50 ± 1.25 | 4: 5 | Genus: Bifidobacterium, Lactobacillus, Prevotella |
| Finegold et al.(2010)[37] | USA | 11 | - | - | 8 | - | - | Phylum: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Fusobacteria, Proteobacteria, Tenericutes |
| Phylum/Genus | Studies included | SMD | 95% CI | I2 | Overall effect(Z) | p value |
|---|---|---|---|---|---|---|
| Actinobacteria | 11 | -0.1 | (-0.46, 0.26) | 87 | 0.53 | 0.60 |
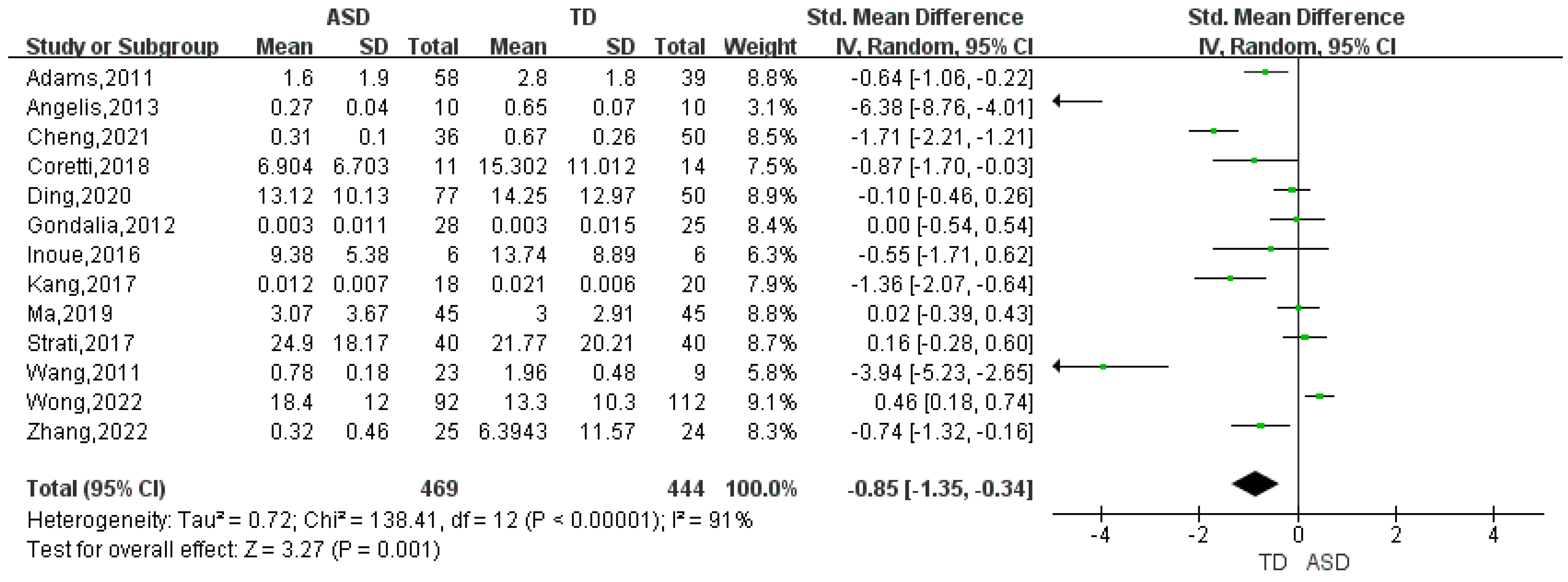
| Bifidobacterium | 13 | -0.85 | (-1.35, -0.34) | 91 | 3.27 | 0.001 |
| Collinsella | 5 | 0.23 | (-0.14, 0.60) | 76 | 1.21 | 0.23 |
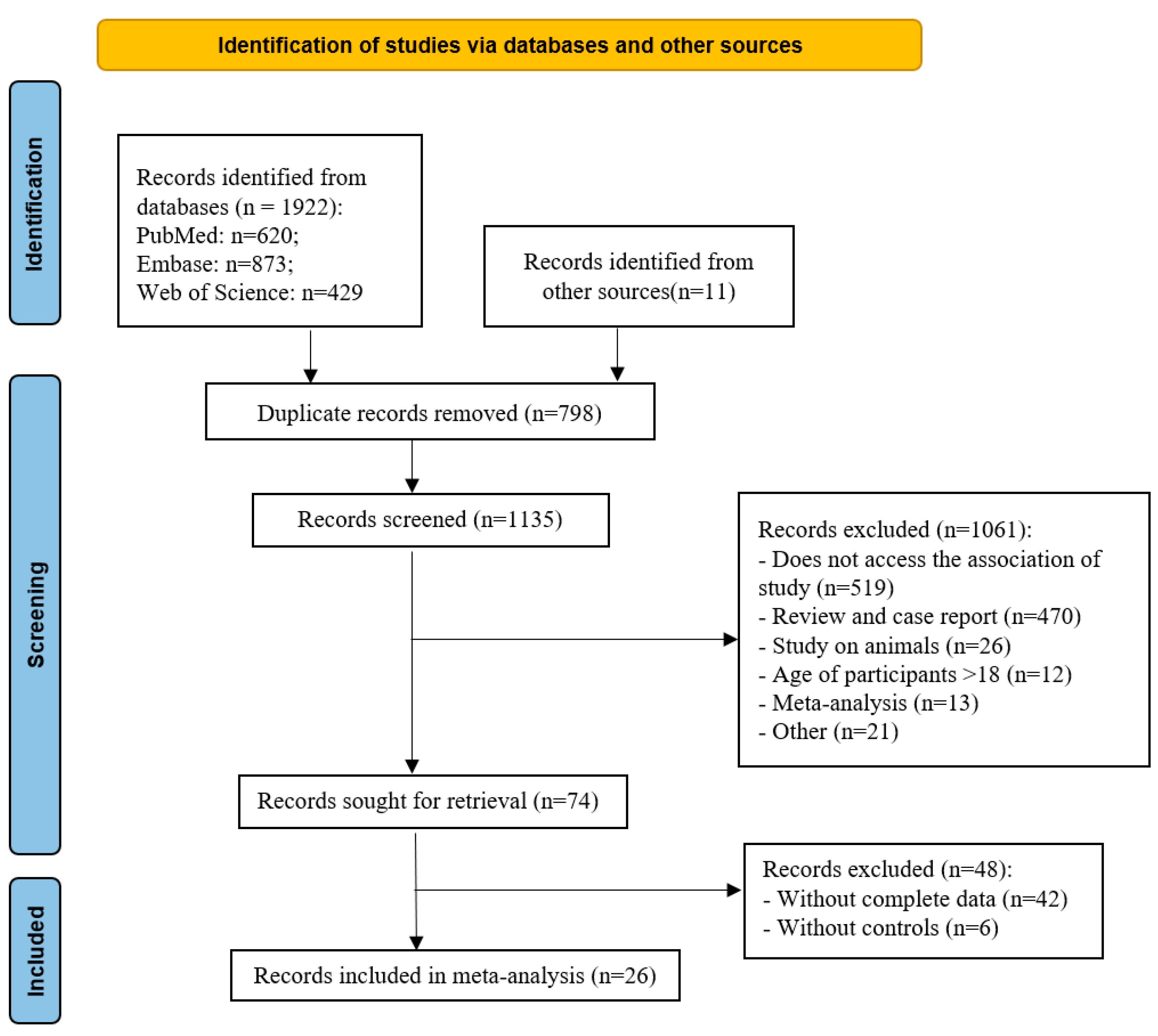
| Bacteroidetes | 12 | 0.42 | (0.02, 0.82) | 89 | 2.05 | 0.04 |
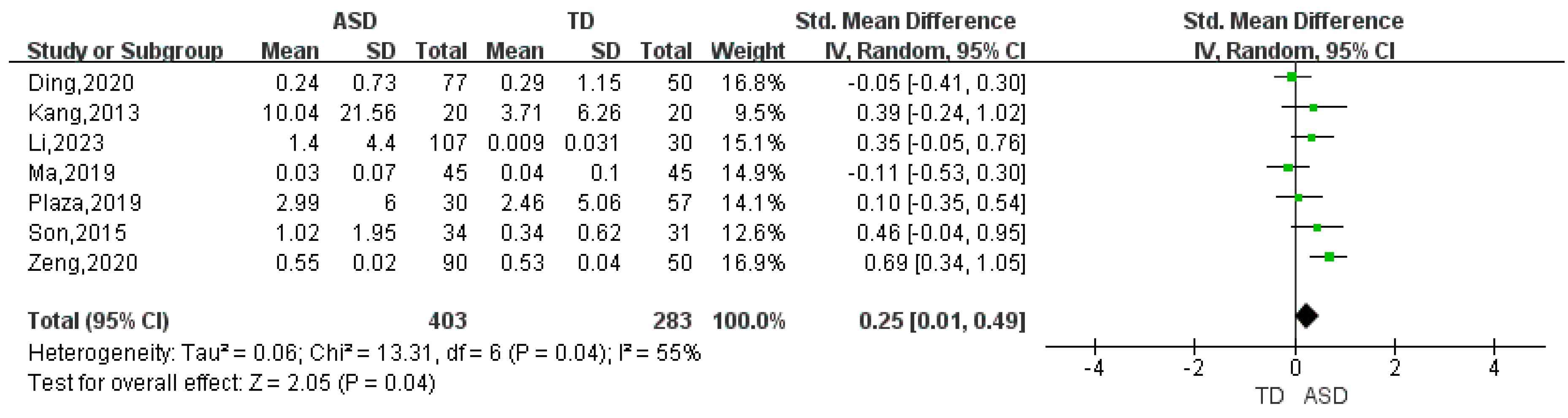
| Bacteroides | 13 | 0.47 | (0.01, 0.93) | 91 | 2.01 | 0.04 |
| Parabacteroides | 6 | -0.05 | (-0.40, 0.31) | 76 | 0.26 | 0.79 |
| Prevotella | 6 | -0.01 | (-0.44, 0.42) | 85 | 0.04 | 0.97 |
| Paraprevotella | 3 | -0.12 | (-0.46, 0.21) | 66 | 0.72 | 0.47 |
| Cyanobacteria | 5 | -0.09 | (-0.59, 0.40) | 86 | 0.38 | 0.71 |
| Firmicutes | 13 | -0.24 | (-0.81, 0.33) | 94 | 0.83 | 0.41 |
| Blautia | 7 | -0.13 | (-0.54, 0.28) | 84 | 0.61 | 0.54 |
| Clostridium | 5 | 1.01 | (0.15, 1.87) | 85 | 2.31 | 0.02 |
| Coprococcus | 7 | -0.43 | (-0.80, -0.06) | 65 | 2.25 | 0.02 |
| Dorea | 4 | 0.5 | (0.31, 0.70) | 0 | 5.03 | <0.001 |
| Faecalibacterium | 8 | -0.02 | (-0.43, 0.40) | 81 | 0.07 | 0.94 |
| Lactobacillus | 4 | 0.27 | (-0.36, 0.89) | 85 | 0.84 | 0.40 |
| Megamonas | 3 | 0.04 | (-0.40, 0.48) | 76 | 0.19 | 0.85 |
| Phascolarctobacterium | 4 | -0.21 | (-0.62, 0.21) | 71 | 0.99 | 0.32 |
| Streptococcus | 4 | -0.47 | (-1.04, 0.10) | 79 | 1.63 | 0.10 |
| Fusobacteria | 6 | -0.1 | (-0.49, 0.28) | 71 | 0.99 | 0.32 |
| Fusobacterium | 3 | 0.05 | (-0.26, 0.36) | 69 | 0.32 | 0.75 |
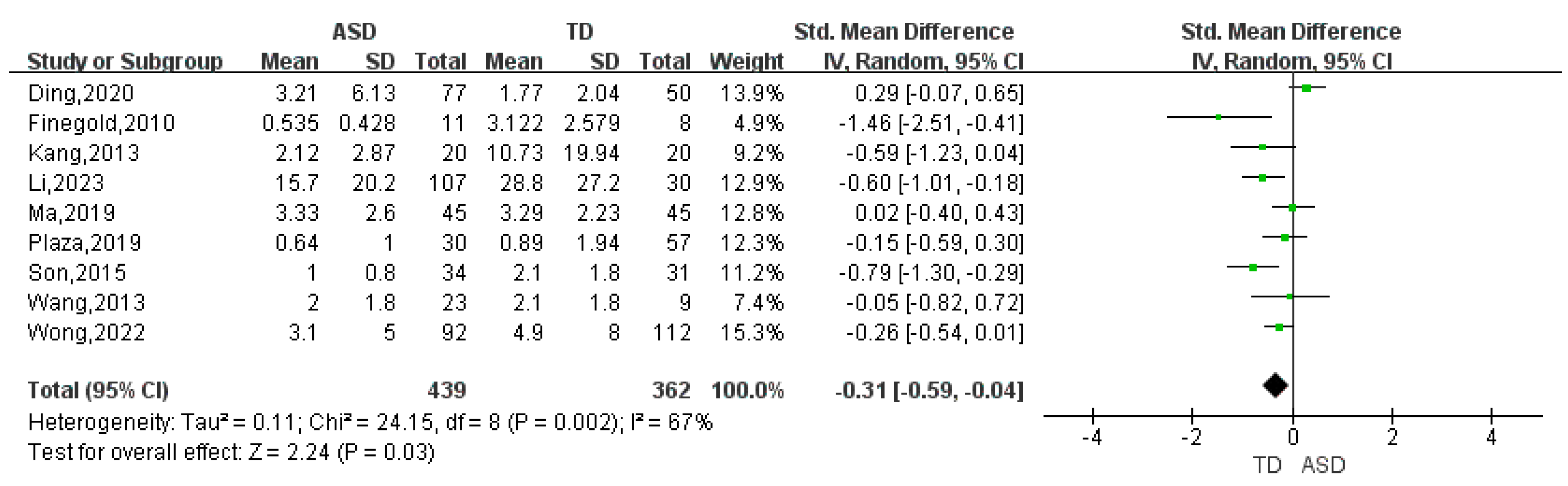
| Proteobacteria | 9 | -0.31 | (-0.59, -0.04) | 67 | 2.24 | 0.03 |
| Bilophila | 3 | -0.36 | (-0.87, 0.15) | 68 | 1.37 | 0.17 |
| Sutterella | 4 | 1.04 | (0.04, 2.05) | 95 | 2.03 | 0.04 |
| Tenericutes | 4 | -0.47 | (-1.08, 0.15) | 84 | 1.48 | 0.14 |
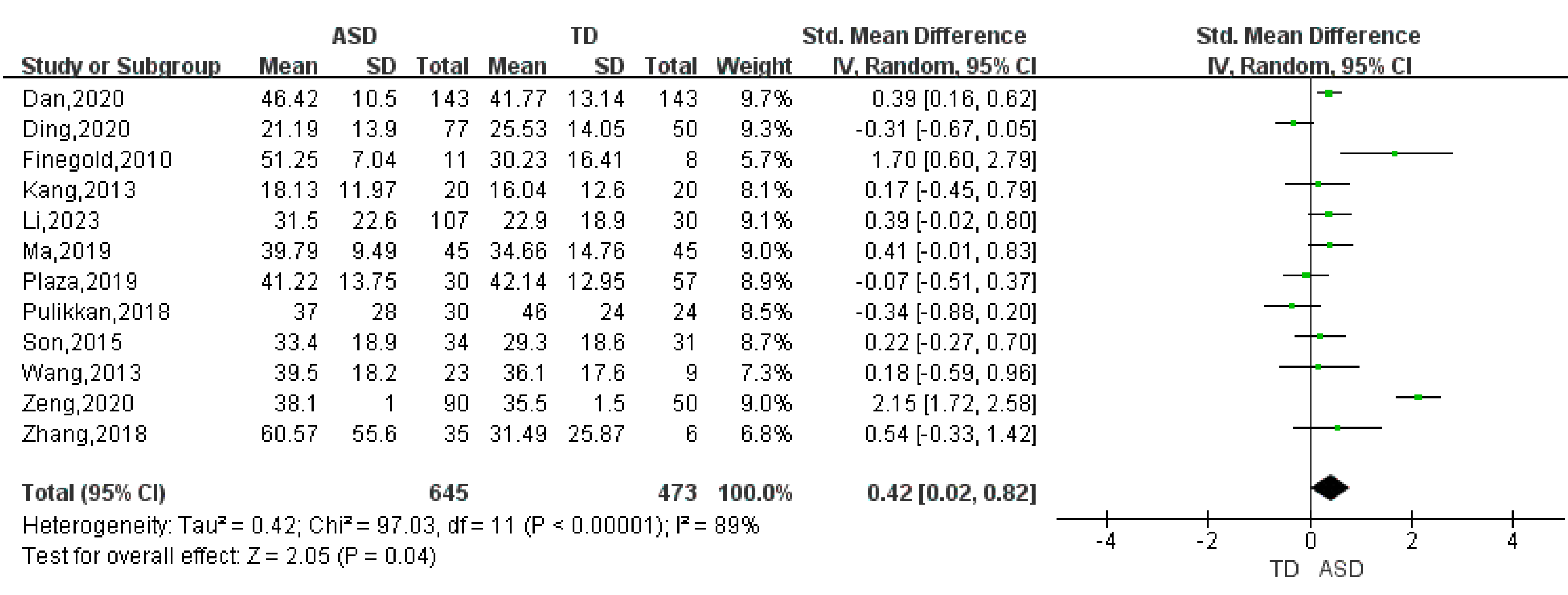
| Verrucomicrobia | 7 | 0.25 | (0.01, 0.49) | 55 | 2.05 | 0.04 |
| Akkermansia | 4 | -0.41 | (-0.77, -0.05) | 38 | 2.26 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




