1. Introduction
The gut microbiota is a complex and diverse community of microorganisms that reside in the gastrointestinal tract [
1]. It plays a crucial role in the maintenance of the host's health and wellbeing by performing important metabolic, immunological, and nutritional functions [
2]. Recent studies have shown that alterations in the gut microbiota composition and diversity can have a significant impact on human health and disease [
3].
The COVID-19 pandemic has brought the importance of gut microbiota into the spotlight, as research has suggested that COVID-19 patients with gastrointestinal symptoms may have a different gut microbiota composition compared to those without such symptoms [
4,
5,
6,
7]. Additionally, individuals with pre-existing conditions such as type 2 diabetes (T2D) have been found to have a less diverse gut microbiota, which may contribute to the progression and severity of COVID-19 [
8].
Antibiotic treatment is commonly used to manage bacterial infections, but it can also affect the gut microbiota by altering its composition and reducing its diversity [
9]. Metformin therapy, a commonly used drug for the treatment of T2D, has also been found to impact the gut microbiota [
10]. Several studies have demonstrated that metformin can increase the abundance of beneficial bacteria, such as
Akkermansia muciniphila and
Faecalibacterium prausnitzii, while reducing the abundance of harmful bacteria such as
Escherichia coli [
11,
12].
In this study, we aimed to investigate the alpha diversity of gut microbiota in patients with COVID-19 and T2D, and to analyze the impact of COVID-19 variants, antibiotic treatment, and metformin therapy on gut microbiota composition and diversity. We used a culture-based method to analyze the gut microbiota, which enabled us to identify and quantify bacterial species present in the gut.
Understanding the impact of these factors on gut microbiota composition and diversity could provide valuable insights into the pathogenesis and management of COVID-19 in patients with T2D, and may lead to the development of targeted interventions aimed at modulating the gut microbiota to improve patient outcomes.
2. Material and Methods
2.1. Study design and sample collection
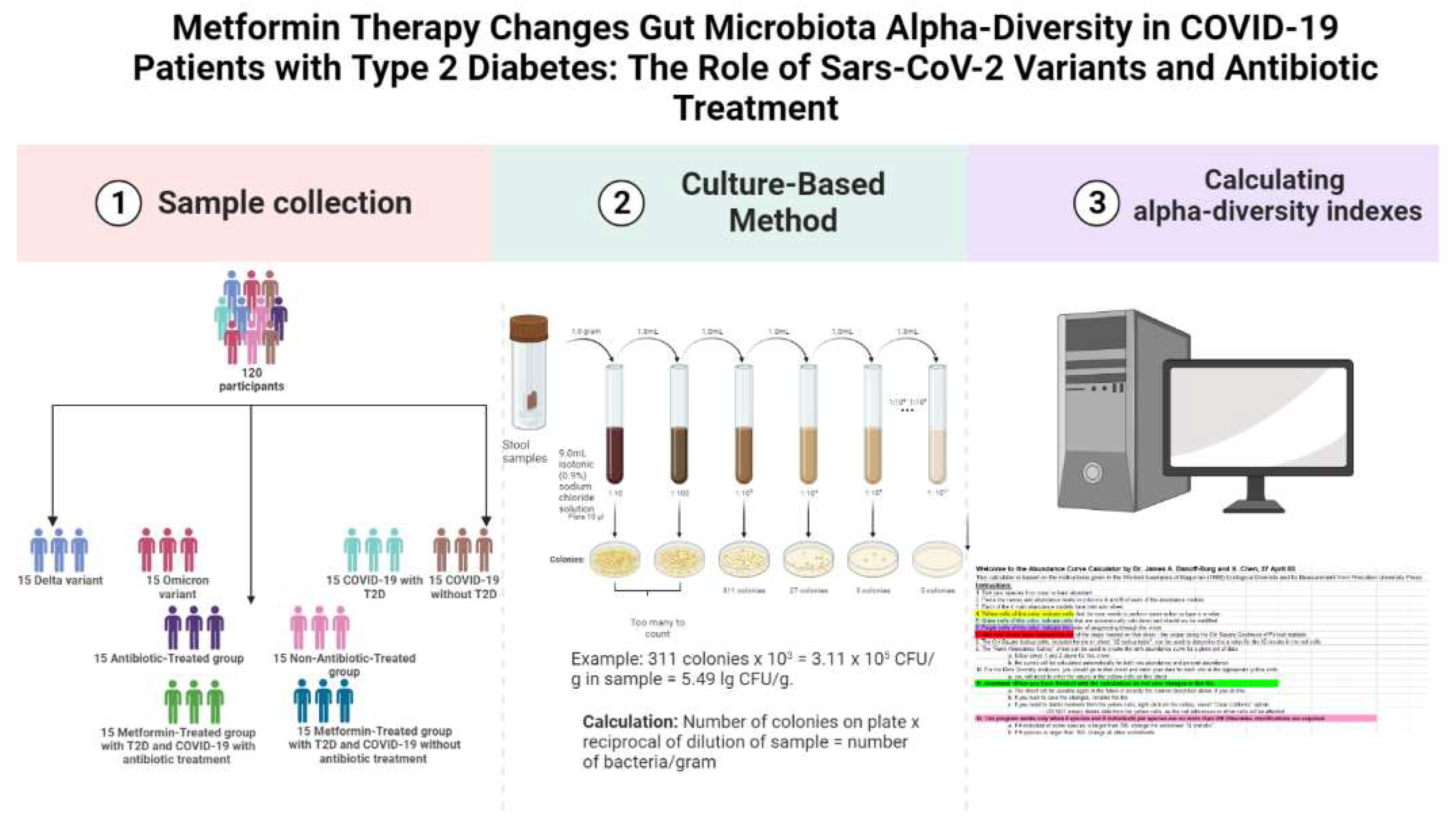
The study was conducted on 120 fecal samples collected from COVID-19 confirmed patients admitted to the Transcarpathian Regional Infectious Hospital from 2020 to 2022 (
Figure 1). The study was conducted in four stages, each analyzing the gut microbiota characteristics of different groups of patients with COVID-19. Patients were assigned to the Delta and Omicron groups based on the predominance of the respective strains during the study period. Additionally, patients were divided into groups based on their antibiotic treatment status, with 29.3% receiving linezolid, 34.4% receiving meropenem, 25.8% receiving fluoroquinolones (moxifloxacin and ciprofloxacin), and 10.5% receiving cephalosporins of the III or IV generations. The Metformin-Treated Patients with T2D (MTP with T2D) group consisted of patients who took metformin in a dose of 1000-1500 mg per day for at least 3 months prior to admission.
2.2. Microbiota analysis and calculation of alpha-diversity
For gut microbiota analysis, the weight of the fecal sample (1.0 gram) was recorded, and 9 ml of isotonic (0.9%) sodium chloride solution was added to a test tube. The mixture was thoroughly rubbed until a homogeneous mass was formed, creating a 10
-1 dilution. Subsequently, a series of dilutions from 10
-2 to 10
-11 were prepared in the same way (
Figure 2). Using sterile micropipettes, 10 μl was taken from each dilution and applied to nutrient media for the isolation of specific microorganisms. Commercial nutrient media was used for the isolation of enterobacteria,
Staphylococcus spp.,
Enterococcus spp., yeast,
Clostridium spp.,
Lactobacillus spp.,
Bifidobacterium spp., and
Bacteroides species (
Supplementary file). Identification of microorganisms was carried out based on the Clinical Microbiology Procedures Handbook, Volume 1-3, 4th Edition [
13]. Decimal logarithms of the quantitative indicator of the grown colonies of microorganisms (lg CFU/g) were used for the convenience of material presentation and mathematical and statistical processing.
To calculate the alpha-diversity of the gut microbiota, we used the Shannon H' and Simpson 1/D indices. The Shannon H' index was calculated using the formula H' = -∑ pi ln(pi), where pi is the proportion of individuals belonging to the ith taxon. The Simpson 1/D index was calculated using the formula 1/D = ∑ pi^2, where pi is the proportion of individuals belonging to the ith taxon. We used the Abundance Curve Calculator by Dr. James A. Danoff-Burg and X. Chen, 27 April 05, to calculate the diversity.
In addition, we collected data on several clinical parameters, including length of hospital stay (LoS), C-reactive protein (CRP) levels, and neutrophil-to-lymphocyte ratio (NLR).
2.3. Statistical analysis
To compare alpha diversity indices between groups, we used a non-parametric test, the Kruskal-Wallis test, followed by Dwass-Steel-Critchlow-Fligner (DSCF) post-hoc test. Spearman rank correlation was used to explore the correlation between LoS, CRP, NLR, and diversity indices. To investigate the relationship between COVID-19 with T2D and COVID-19 without T2D, we used binary logistic regression. The independent variables included in the model were the alpha diversity indices. We calculated odds ratios and 95% confidence intervals, and set statistical significance at p < 0.05.
3. Results
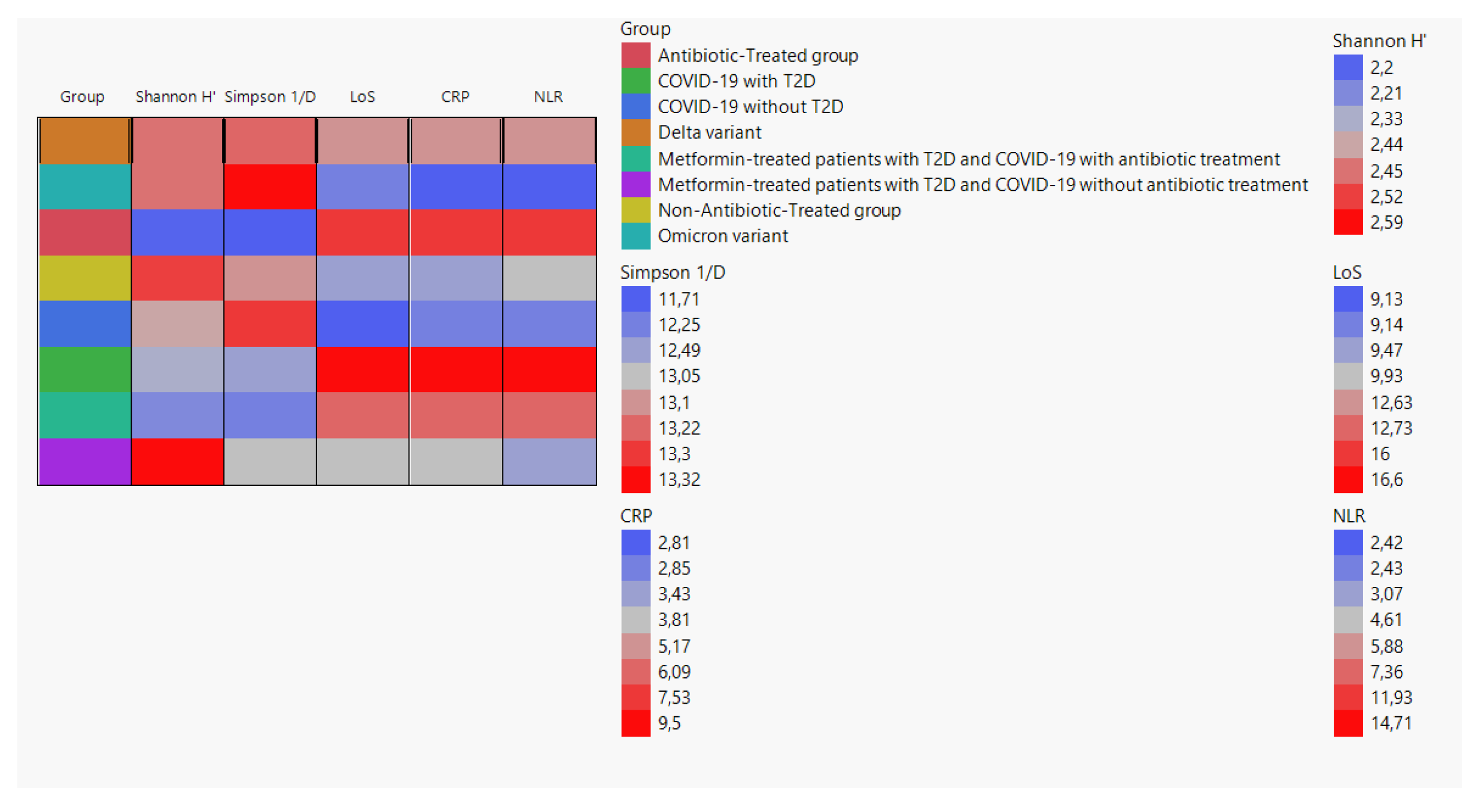
We created a cell plot to visualize the average values of Shannon H', Simpson 1/D, LoS, CRP, and NLR among different groups. The data used for the cell plot was obtained from the following groups: Delta variant, Omicron variant, antibiotic-treated group, non-antibiotic-treated group, COVID-19 without T2D, COVID-19 with T2D, metformin-treated patients with T2D and COVID-19 with antibiotic treatment, and metformin-treated patients with T2D and COVID-19 without antibiotic treatment (
Figure 2).
3.1. Alpha-diversity analysis
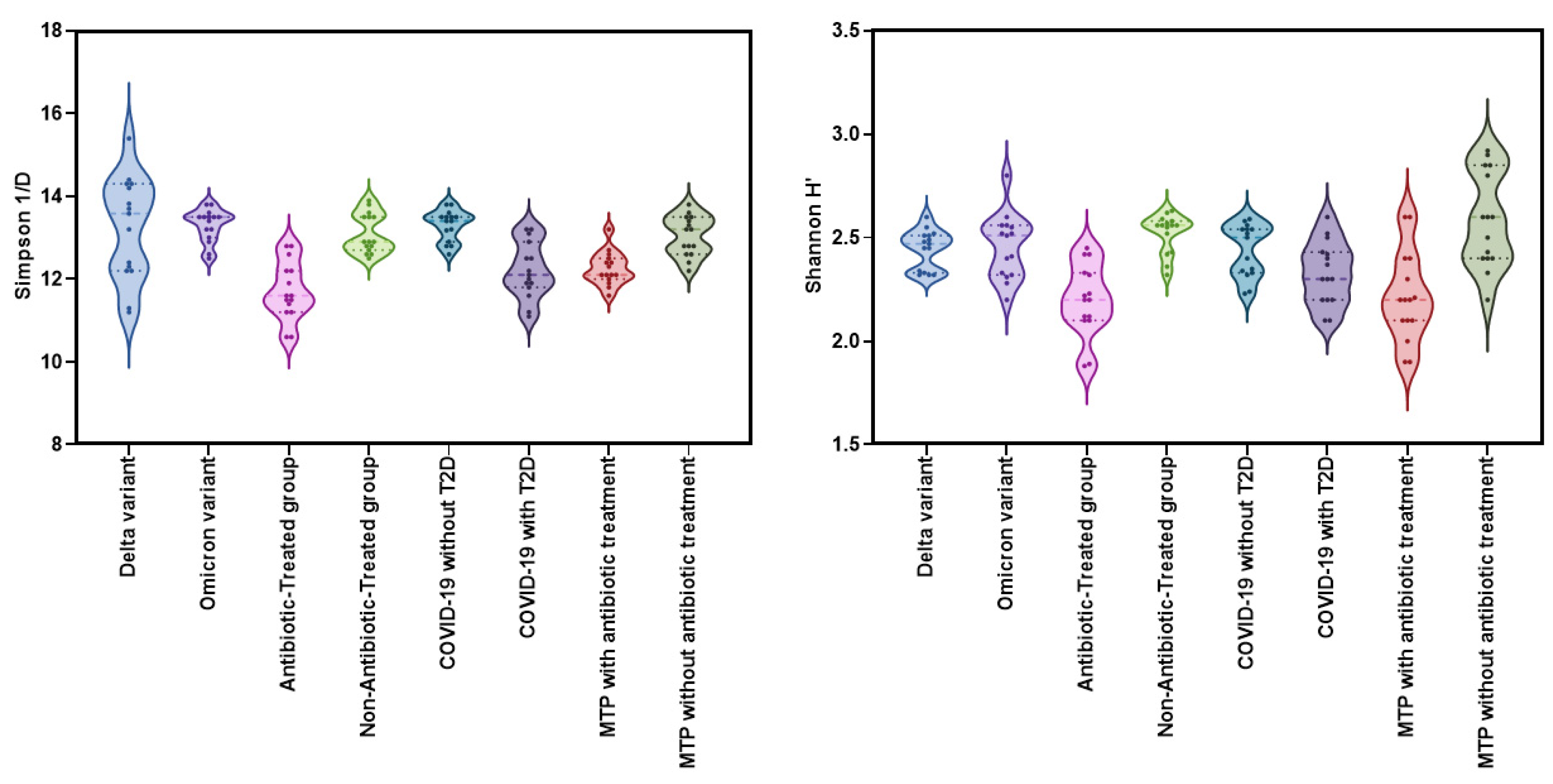
The Kruskal-Wallis test was used to compare the alpha diversity indices (Shannon H' and Simpson 1/D) among eight groups: Delta variant, Omicron variant, COVID-19 without T2D, COVID-19 with T2D, Metformin-treated patients with T2D and COVID-19 with antibiotic treatment, Metformin-treated patients with T2D and COVID-19 without antibiotic treatment, Antibiotic-Treated group, and Non-Antibiotic-Treated group. The results showed significant differences among the groups for both Shannon H' (χ²=45.3, df=9, p<0.001, ε²=0.381) and Simpson 1/D (χ²=57.3, df=9, p<0.001, ε²=0.482) (
Figure 3).
The Shannon H index did not show any significant differences in alpha-diversity between the Delta and Omicron variant groups (W=0.588; p=1.000). The Antibiotic-Treated group had significantly lower diversity (W=5.995; p<0.001) compared to the Non-Antibiotic-Treated group. However, there was no significant difference in the Shannon H index (p=0.544) between COVID-19 patients with and without T2D, suggesting that the presence of T2D may not significantly affect alpha-diversity in patients with COVID-19 and T2D.
Significant differences were observed in Shannon H' index (W=5.017, p=0.014) between Metformin-treated patients with T2D and COVID-19 with antibiotic treatment and Metformin-treated patients with T2D and COVID-19 without antibiotic treatment. The DSCF test revealed a significant difference in Simpson 1/d (W=5.913; p<0.001) between the Antibiotic-Treated group and the Non-Antibiotic-Treated group. The DSCF test also indicated a significant difference in Simpson 1/d (p=0.006) between Metformin-treated patients with T2D and COVID-19 with antibiotic treatment and Metformin-treated patients with T2D and COVID-19 without antibiotic treatment. Furthermore, significant differences in Simpson 1/d were observed between COVID-19 patients without T2D and those with T2D (W=-5.352; p=0.006) (
Figure 4).
3.2. Correlation between Alpha Diversity Indices and Clinical Parameters
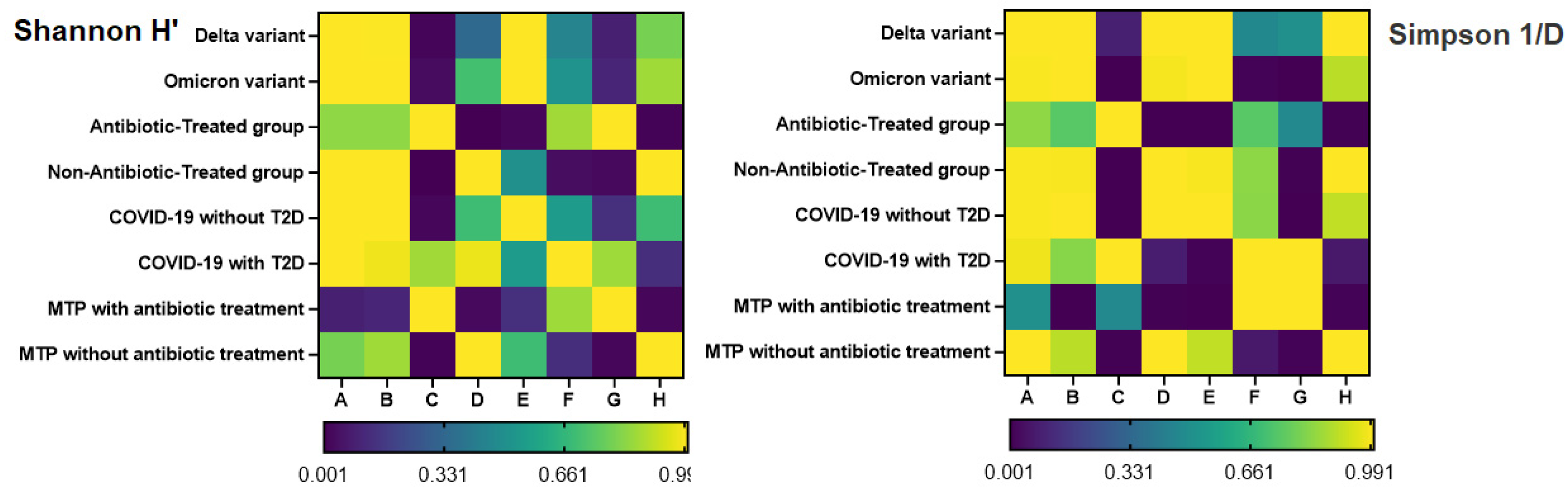
In our study, we conducted a correlation analysis between alpha diversity indices (Shannon H' and Simpson 1/D), LoS, CRP, and NLR in our patient population. Spearman rank correlation was used to analyze the data, and the results are presented in the correlation matrix.
The Shannon H' index showed a significant positive correlation with the Simpson 1/D index (Spearman's rho = 0.717, p < 0.001). Additionally, there was a significant negative correlation between the Shannon H' index and all three clinical variables: LoS (Spearman's rho = -0.563, p < 0.001), CRP (r = -0.553, p < 0.001), and NLR (r = -0.519, p < 0.001). The Simpson 1/D index showed a similar negative correlation with LoS, CRP, and NLR (Spearman's rho ranged from -0.727 to -0.748, all p < 0.001) (
Figure 5).
These findings suggest that decreased alpha diversity is associated with increased LoS, higher levels of CRP, and a higher NLR in our patient population. These correlations may provide insight into the underlying mechanisms and clinical implications of altered gut microbiota in COVID-19 patients.
3.3. Binary logistic regression
A predictive model was developed to estimate the probability of COVID-19 with T2D, conditioning on Shannon H' and Simpson 1/D, using binary logistic regression.
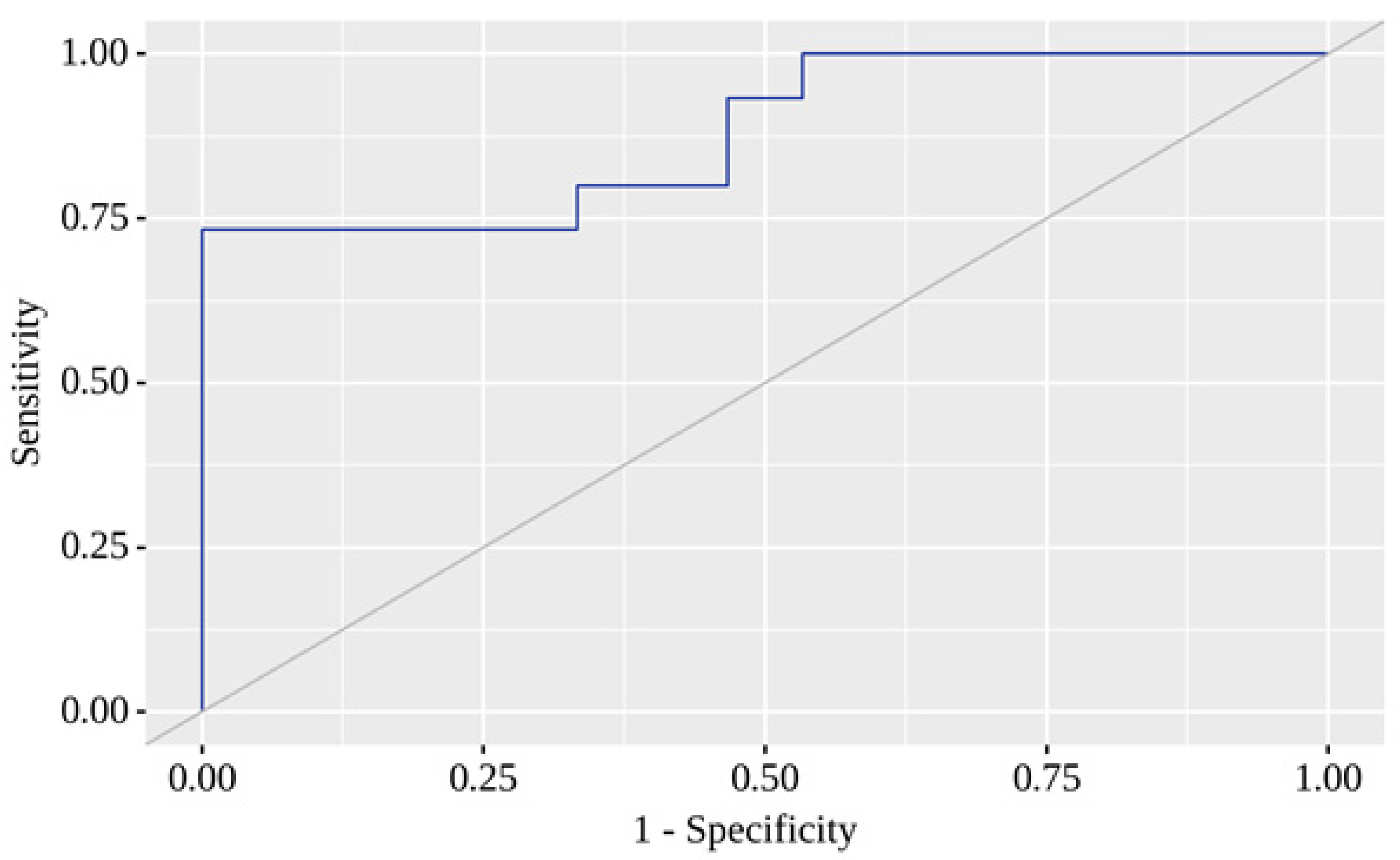
Here, P represents the probability of a positive result. The resulting regression model was statistically significant (p < 0.001). Based on the Nagelkerke R² value, the model explains 59.1% of the observed COVID-19 with T2D variance. When evaluating the dependence of the probability of a positive result on the value of the logistic function P using ROC analysis, the resulting curve had an area under the ROC curve of 0.880 ± 0.065 with 95% CI: 0.752 - 1.000 (
Figure 6).
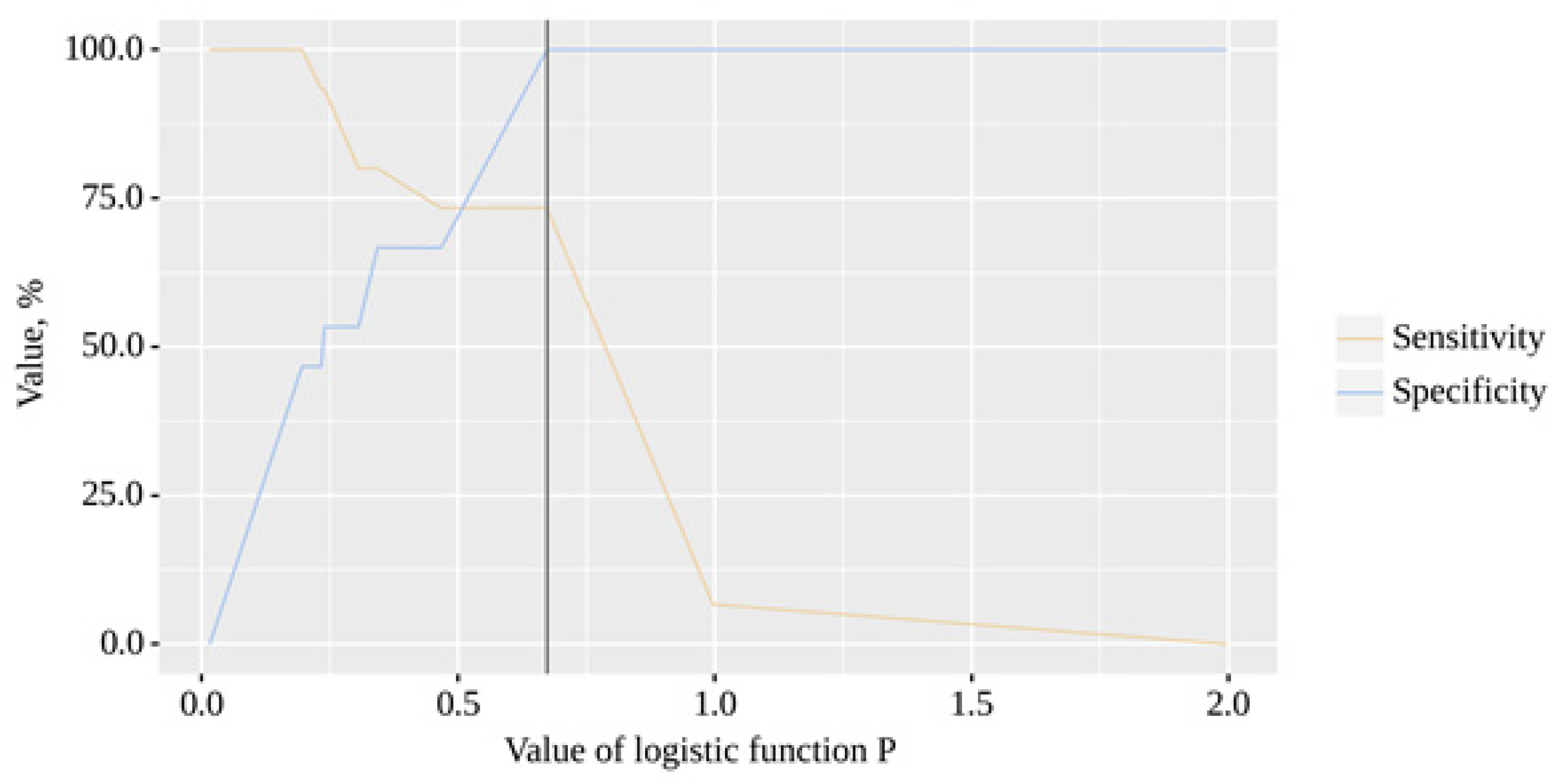
The resulting model was statistically significant (p < 0.001). The cut-off value of the logistic function P, which corresponds to the highest Youden's J statistic, is 0.673. If the value of the logistic function P was greater than or equal to this value, a positive result was predicted. The sensitivity and specificity of the method were 73.3% and 100.0%, respectively (
Figure 7).
4. Discussion
In this study, we investigated alterations in gut microbiota diversity in patients with COVID-19 and T2D, as well as the potential association between gut microbiota diversity and clinical parameters. Our findings indicate that the use of antibiotics and the presence of T2D in COVID-19 patients may have an impact on gut microbiota diversity, as demonstrated by the significant differences in alpha diversity indices observed among the eight groups. Specifically, the Antibiotic-Treated group had significantly lower diversity compared to the Non-Antibiotic-Treated group, and there was no significant difference in the Shannon H index between COVID-19 without T2D and COVID-19 with T2D, suggesting that T2D may not significantly affect alpha diversity in COVID-19 patients with T2D. The findings from our study reveal that there are significant differences in alpha-diversity (Shannon H) between Metformin-treated patients with T2D and COVID-19 with antibiotic treatment and Metformin-treated patients with T2D and COVID-19 without antibiotic treatment. Specifically, the alpha-diversity was found to be higher in the group of patients who did not receive antibiotic treatment compared to those who did.
This observation is of significant interest as it suggests that the use of antibiotics may have a detrimental effect on the gut microbiome in individuals with T2D and COVID-19 who are being treated with Metformin. Previous studies have demonstrated the importance of a healthy gut microbiome in regulating glucose metabolism and maintaining overall metabolic health in individuals with T2D. Therefore, the observed differences in alpha-diversity between the two groups of patients may have important clinical implications. Our findings are consistent with those reported by other researchers who have investigated the impact of antibiotics on gut microbiota diversity in various patient populations, including COVID-19 patients [
14,
15,
16,
17,
18].
Our study also revealed a significant positive correlation between the Shannon H' and Simpson 1/D indices and a significant negative correlation between these indices and clinical parameters such as LoS, CRP, and NLR in our patient population. These findings suggest that decreased alpha diversity is associated with increased LoS, higher levels of CRP, and a higher NLR in our patient population. This is in line with previous studies that have reported similar correlations between gut microbiota and clinical parameters such as disease severity, inflammation, and immune response in various patient populations, including COVID-19 patients [
8,
19]. Furthermore, our predictive model, which estimates the probability of COVID-19 with T2D based on Shannon H' and Simpson 1/D, demonstrated good predictive accuracy, with an area under the ROC curve of 0.880.
Although our study contributes to the growing body of literature on the impact of COVID-19 and T2D on gut microbiota diversity and the potential association between gut microbiota diversity and clinical parameters, there are some limitations to our study. First, our sample size was relatively small, which may limit the generalizability of our findings. Second, it is worth noting that the observed differences in alpha-diversity may be influenced by several factors such as diet, age, and disease severity, which were not controlled for in our study. Moreover, the study was limited by lack of a control group of patients with T2D who did not have COVID-19.
In conclusion, our study provides evidence for the potential impact of antibiotics and T2D on gut microbiota diversity in COVID-19 patients and the potential association between gut microbiota diversity and clinical parameters. Our findings may have implications for the development of personalized therapeutic strategies for COVID-19 patients with T2D that target the gut microbiota [
20,
21,
22,
23,
24,
25].
Author Contributions
Conceptualization, A.K.; methodology, A.K. and P.P..; software, A.K. and P.P.; validation, A.K., P.P. and I.K.; formal analysis, I.K.; investigation, P.P.; resources, A.K.; data curation, A.K.; writing—original draft preparation, P.P.; writing—review and editing, A.K., V.O., D.K.; visualization, P.P. and A.K.; supervision, A.K.; project administration, A.K.. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Uzhhorod National Univesrity (11.02.20/11ab).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thursby E, Juge N. Introduction to the human gut microbiota. The Biochemical journal. 2017;474(11):1823-36. [CrossRef]
- Valentini M, Piermattei A, Di Sante G, Migliara G, Delogu G, Ria F. Immunomodulation by Gut Microbiota: Role of Toll-Like Receptor Expressed by T Cells. Journal of Immunology Research. 2014;2014:586939. [CrossRef]
- Hou K, Wu Z-X, Chen X-Y, Wang J-Q, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduction and Targeted Therapy. 2022;7(1):135.
- Hussain I, Cher GLY, Abid MA, Abid MB. Role of Gut Microbiome in COVID-19: An Insight Into Pathogenesis and Therapeutic Potential. Front Immunol. 2021;12:765965. [CrossRef]
- Petakh P, Isevych V, Mohammed I, Loshak K, Poliak I, Kamyshnyiy A. Association between Use of Metformin and Insulin with Hematological Parameters in COVID-19 Patients with Type 2 Diabetes: A Single Center, Cross-Sectional Study. Clinical Diabetology. 2022;11(6):432-3. [CrossRef]
- Petakh P, Griga V, Mohammed IB, Loshak K, Poliak I, Kamyshnyiy A. Effects of Metformin, Insulin on Hematological Parameters of COVID-19 Patients with Type 2 Diabetes. Medical archives (Sarajevo, Bosnia and Herzegovina). 2022;76(5):329-32. [CrossRef]
- Petakh P, Kamyshna I, Nykyforuk A, Yao R, Imbery JF, Oksenych V, et al. Immunoregulatory Intestinal Microbiota and COVID-19 in Patients with Type Two Diabetes: A Double-Edged Sword. J Viruses. 2022;14(3):477. [CrossRef]
- Gradisteanu Pircalabioru G, Grigore GA, Czobor Barbu I, Chifiriuc M-C, Savu O. Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines. 2023;11(1):179.
- Patangia DV, Anthony Ryan C, Dempsey E, Paul Ross R, Stanton C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen. 2022;11(1):e1260. [CrossRef]
- Zhang Q, Hu NJD, Metabolic Syndrome, Targets O, Therapy. Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. 2020;13:5003.
- de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care. 2017;40(1):54-62. [CrossRef]
- Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nature medicine. 2017;23(7):850-8. [CrossRef]
- Biochemical Tests for the Identification of Aerobic Bacteria. Clinical Microbiology Procedures Handbook2016. p. 3.17.1.1-3..48.3.
- Bernard-Raichon L, Venzon M, Klein J, Axelrad JE, Zhang C, Sullivan AP, et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nature Communications. 2022;13(1):5926. [CrossRef]
- Righi E, Lambertenghi L, Gorska A, Sciammarella C, Ivaldi F, Mirandola M, et al. Impact of COVID-19 and Antibiotic Treatments on Gut Microbiome: A Role for Enterococcus spp. J Biomedicines. 2022;10(11):2786. [CrossRef]
- Romani L, Del Chierico F, Macari G, Pane S, Ristori MV, Guarrasi V, et al. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front Cell Infect Microbiol. 2022;12:908492.
- Yin YS, Minacapelli CD, Parmar V, Catalano CC, Bhurwal A, Gupta K, et al. Alterations of the fecal microbiota in relation to acute COVID-19 infection and recovery. Molecular biomedicine. 2022;3(1):36. [CrossRef]
- Rebelo JS, Domingues CPF, Dionisio F, Gomes MC, Botelho A, Nogueira T. COVID-19 Lockdowns May Reduce Resistance Genes Diversity in the Human Microbiome and the Need for Antibiotics. International journal of molecular sciences. 2021;22(13). [CrossRef]
- Petakh P, Kobyliak N, Kamyshnyi A. Gut microbiota in patients with COVID-19 and type 2 diabetes: A culture-based method. 2023;13.
- Kamyshnyi O, Matskevych V, Lenchuk T, Strilbytska O, Storey K, Lushchak O. Metformin to decrease COVID-19 severity and mortality: Molecular mechanisms and therapeutic potential. Biomedicine & Pharmacotherapy. 2021;144:112230. [CrossRef]
- Buchynskyi M, Kamyshna I, Lyubomirskaya K, Moshynets O, Kobyliak N, Oksenych V, et al. Efficacy of interferon alpha for the treatment of hospitalized patients with COVID-19: A meta-analysis. 2023;14. [CrossRef]
- Kamyshnyi A, Koval H, Kobevko O, Buchynskyi M, Oksenych V, Kainov D, et al. Therapeutic Effectiveness of Interferon-α2b against COVID-19 with Community-Acquired Pneumonia: The Ukrainian Experience. 2023;24(8):6887. [CrossRef]
- Ianevski A, Ahmad S, Anunnitipat K, Oksenych V, Zusinaite E, Tenson T, et al. Seven classes of antiviral agents. Cellular and Molecular Life Sciences. 2022;79(12):605.
- Ianevski A, Simonsen RM, Myhre V, Tenson T, Oksenych V, Bjørås M, et al. DrugVirus.info 2.0: an integrative data portal for broad-spectrum antivirals (BSA) and BSA-containing drug combinations (BCCs). Nucleic acids research. 2022;50(W1):W272-w5. [CrossRef]
- Ianevski A, Yao R, Simonsen RM, Myhre V, Ravlo E, Kaynova GD, et al. Mono- and combinational drug therapies for global viral pandemic preparedness. iScience. 2022;25(4):104112. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).