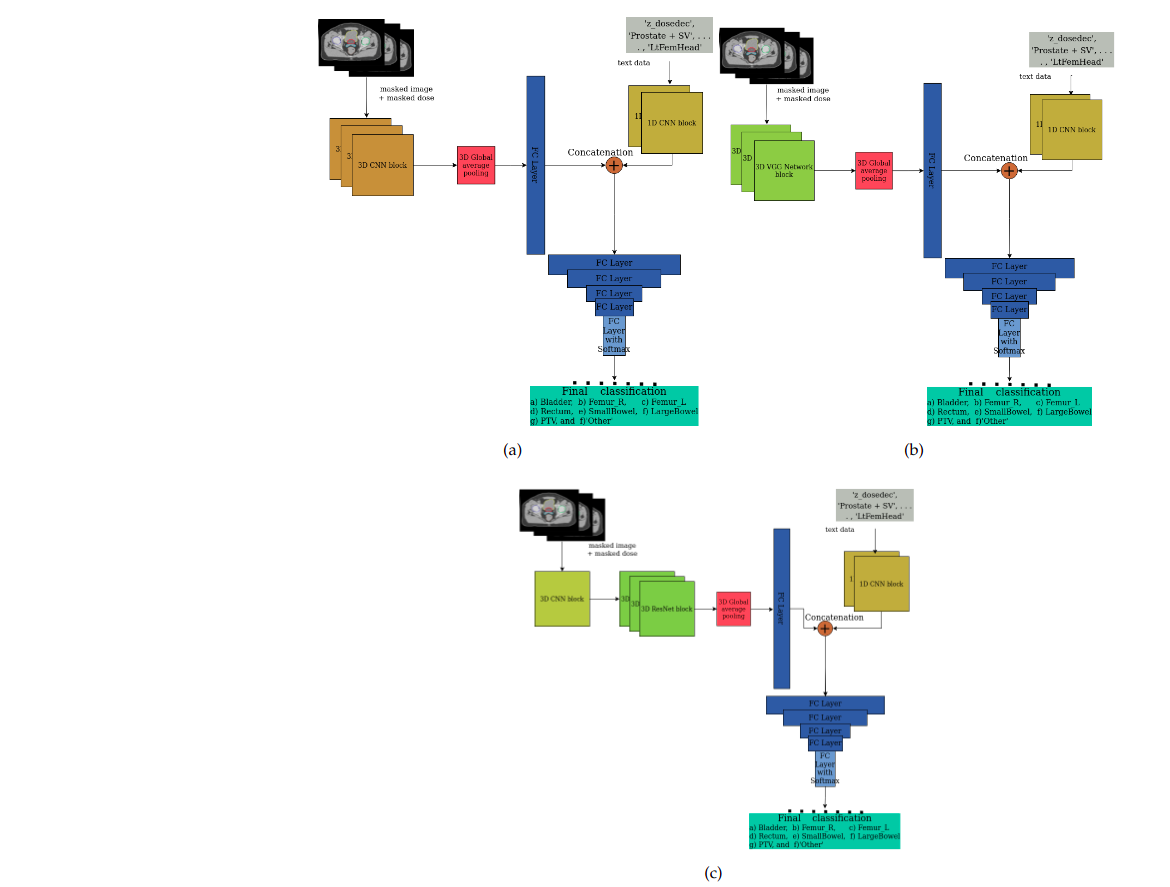
Physicians often label anatomical structure sets in Digital Imaging and Communications in Medicine (DICOM) images with nonstandard names. As these names vary widely, the standardization of the nonstandard names in the Organs at Risk (OARs), Planning Target Volumes (PTVs), and 'Other' organs inside the area of interest is a vital problem. Prior works considered traditional machine learning approaches on structure sets with moderate success. This paper presents integrated deep learning methods applied to structure sets by integrating the multimodal data compiled from the radiotherapy centers administered by the US Veterans Health Administration (VHA) and the Department of Radiation Oncology at Virginia Commonwealth University (VCU). The de-identified radiation oncology data collected from VHA and VCU radiotherapy centers have 16,290 prostate structures. Our method integrates the heterogeneous (textual and imaging) multimodal data with Convolutional Neural Network (CNN)-based deep learning approaches like CNN, Visual Geometry Group (VGG) network, and Residual Network (ResNet). Our model presents improved results in prostate (RT) structure name standardization. Evaluation of our methods with macro-averaged F1 Score shows that our deep learning model with single-modal textual data usually performs better than the previous studies. We also experimented with various combinations of multimodal data (masked images, masked dose) besides textual data. The models perform well on the textual data alone, while the addition of imaging data shows that deep neural networks can achieve improved performance using information present in the other modalities. Additionally, using masked images and masked doses along with text leads to an overall performance improvement with the various CNN-based architectures than using all the modalities together. Undersampling the majority class leads to further performance enhancement. The VGG network on the masked image-dose data combined with CNNs on the text data performs the best and establishes the state-of-the-art in this domain.