1. Introduction
From the late 1940s to the 1970s, many antibiotic molecules of natural or synthetic origin were discovered. The dazzling success of the first anti-infective treatments caused the problem of infectious diseases to be considered somewhat hastily as definitively settled. However, the enthusiasm quickly waned with the appearance of the first bacterial resistance to antibiotics [
1]. With each new antibiotic introduced into therapy, the bacteria have been able to adapt and resist more or less quickly [
2]. Today, bacteria are resistant to all potentially active antibiotics, although antibiotic resistance can be natural or acquired [
3]. Natural resistance is expressed by all strains of a species and its genetic support is a mobile genetic element while acquired resistance is present only in certain strains within a given species. This resistance may be due to mutations affecting genes present on the chromosome or to the acquisition of foreign genes [
3]. These genes can come from the chromosome of different species or be conveyed by mobile genetic elements that can be transferred from one bacterium to another, we speak of horizontal transmission because it occurs outside of any mechanism of reproduction. This transfer allows rapid diffusion of resistance genes and can sometimes take place between very distant bacteria on the phylogenetic level, or even between Gram-negative bacteria and Gram-positive bacteria [
4]. According to epidemiological data, the appearance and increase of bacterial resistance concerns many countries, with variations depending on the type of microorganism and depending on the country. They concern bacterial species with an important role in community infectious diseases [
4].
In February 2017, the World Health Organization (WHO) published its first list of priority pathogens resistant to antibiotics, thus requiring the research and development of new molecules [
5]. The list in question enumerates the 12 families of bacteria most threatening to human health. This work by WHO is part of its efforts to combat growing antimicrobial resistance around the world [
5]. It is a new tool to ensure that research and development meets urgent public health needs. The most critical group includes multi-drug resistant (BMR) bacteria that pose a particular threat in hospitals, or for patients whose care requires the use of devices such as respirators or blood catheters [
5]. It includes Acinetobacter producing a carbapenemase,
Pseudomonas producing a carbapenemase and various
enterobacteriaceae resistant to carbapenems and beta-lactams [
5]. They can cause severe, often fatal infections, such as blood infections and pneumonia. The second and third groups on the list (the high and medium priority categories) include other increasingly resistant bacteria causing more common illnesses such as gonorrhea or
salmonella food poisoning. In more detail we find in Priority 2 (high):
Enterococcus faecium (resistance to vancomycin);
Staphylococcus aureus (methicylline resistance, intermediate or complete vancomycin resistance);
Helicobacter pylori (resistance to clarithromycin);
Campylobacter spp (resistance to fluoroquinolones);
Salmonellae (resistance to fluoroquinolones);
Neisseria gonorrhoeae, (cephalosporin resistance, fluoroquinolone resistance) [
5]. In priority 3 (medium):
Streptococcus pneumoniae (insensitive to penicillin);
Haemophilus influenzae (ampicillin resistance);
Shigella spp. (resistant to fluoroquinolones). These multi-resistant bacteria (BMR) are isolated either during the diagnosis of pathological samples or during screening, which identifies patients colonized by BMR and reduces the frequency of their isolation [
5]. The problem is that most of the bacteria that have become resistant or multi-resistant are human commensal bacteria, benefiting from the selection pressure of antibiotics whose frequency of use is high and regular [
4]. This dissemination of multi-resistant bacteria concerns bacterial species having an important role in community infectiology such as
Streptococcus pneumoniae and in nosocomial infectiology like
Staphylococcus aureus,
Enterobacteriaceae,
Acinetobacter baumanii and
Pseudomonas aeruginosa. Bacteria are talked to be multi-resistant to antibiotics (BMR) when, due to the accumulation of acquired resistance to more than 03 families of antibiotics, they are no longer sensitive to more than a small number of antibiotics that can be used therapeutically [
4]. The high frequency, the pathogenic potential and the commensal nature expose BMRs to the risk of their dissemination outside the hospital. The clonal and easily transferable nature of the mobile genetic element allows these BMRs to transfer several resistance factors to other bacteria, which will be expressed in this new bacterial host. This is called "co-resistance". BMRs pose a major therapeutic problem in terms of management and prognosis, and therefore in general public health [
6].
The COVID-19 pandemic has considerably modified the medical activities within the health structures as well as the behavior of the populations, with regard to the use of probabilistic antibiotic therapy on a recurrent basis [
7]. Faced with the epidemic peak of respiratory distress and the adoption of prophylaxis against opportunistic COVID-19 infections, the use of certain antibiotics, in particular Cefixime and Azythromycin, have increased considerably to the point of arriving at a situation of shortage of supply [
7]. Antibiotic consumption increased by 19% compared to the previous year over the period March-April 2019 (768 against 644 DDD/1000 DHD) [
7]. The most prescribed antibiotics and the most important in consumption during the onset of the pandemic were: Amoxicillin (+7%), Cefixime (+30%), Imipenem (+66%), Azythromycin and Erythromycin (+138% each) [
7]. During the occurrence of this pandemic, prescriptions or numerous, prolonged self-medications with an increasingly broad spectrum of antibiotics, even the use of last-line molecules for rescue were significantly increased [
7]. Antibiotic prophylaxis administered to palliate opportunistic bacterial infections in patients with suspected COVID-19 infection probably influenced the selection pressure of commensal bacteria in this pandemic context[
7]. It therefore sounds really interesting to investigate the resistance of bacteria to antibiotics after the COVID-19 pandemics era. The aim of this study was to determine the prevalence and antibiotic resistance pattern of bacteria isolated in 02 referral health facilities in Yaounde before and during the COVID-19 pandemic era.
3. Discussion
In order to determine the prevalence and antibiotic resistance pattern of bacteria isolated in 02 referral health facilities in Yaounde before and during the COVID-19 pandemic era, a retrospective study was conducted in 02 referral health facilities in Yaounde, Cameroon. For that purpose, data were collected from each hospital databases (registries, laboratory books) before (2019) and during the pandemic (2020 and 2021), then, analyzed thereafter.
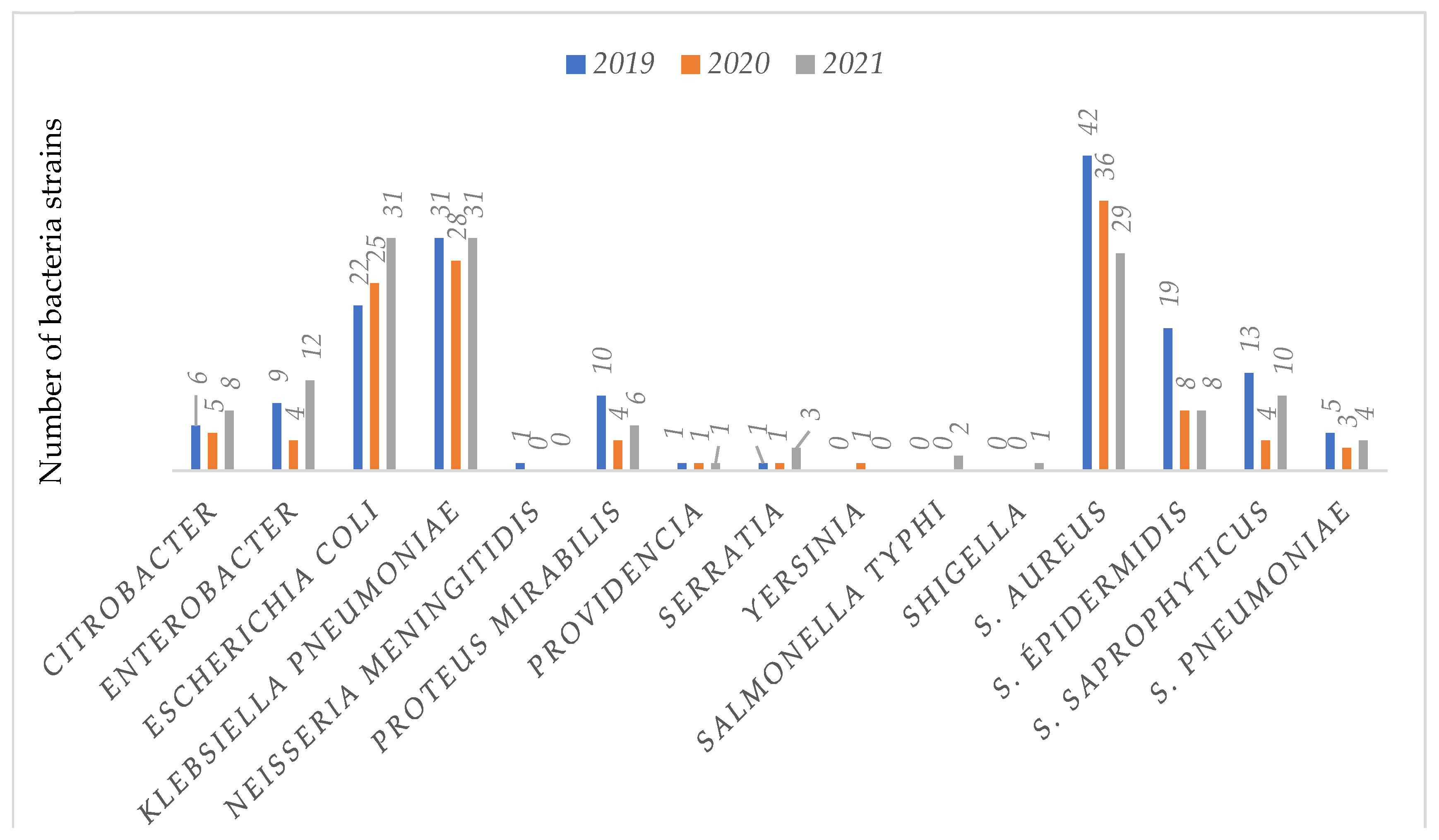
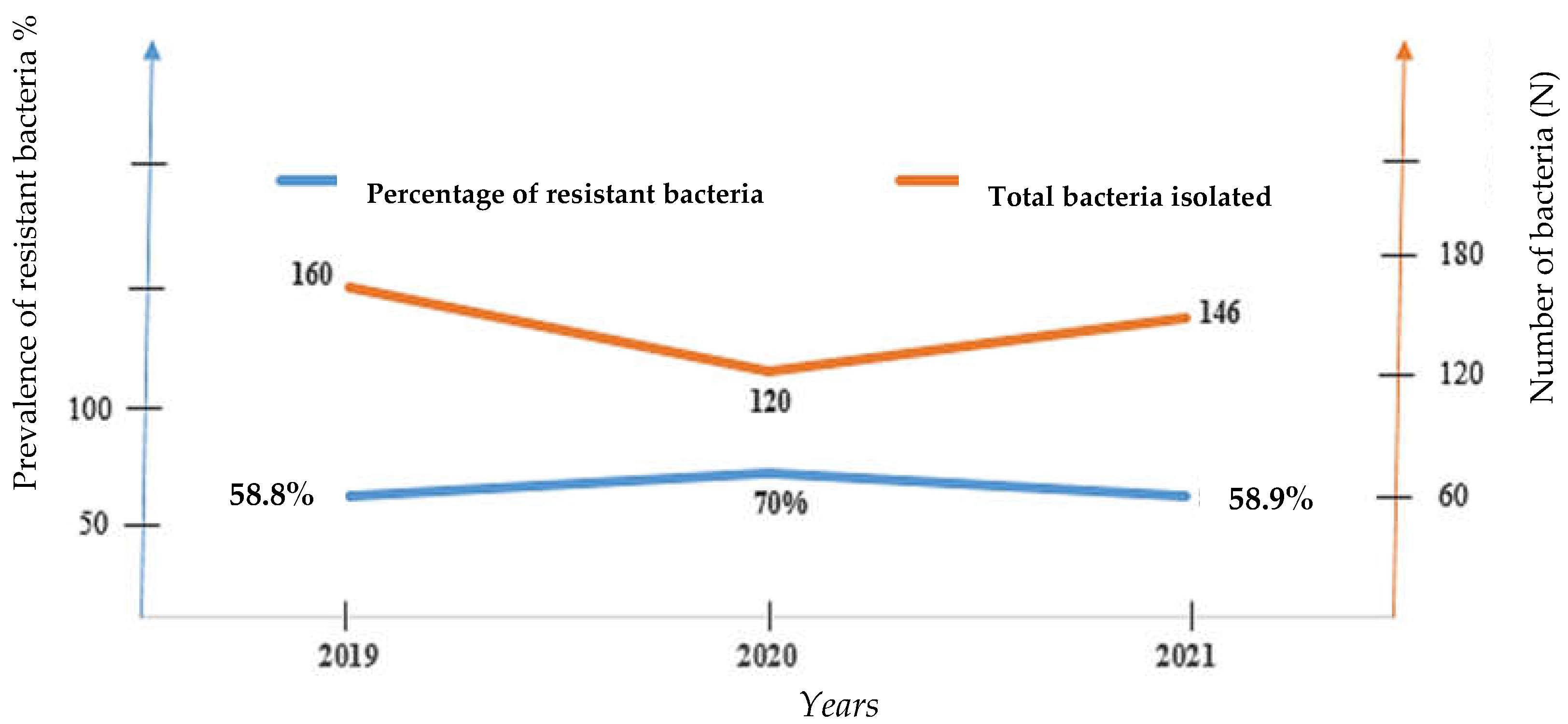
In all, 426 strains were isolated from patients attending the bacteriology unit (a single bacterial strain per patient) of the 02 referral hospitals for COVID-19 as follow: 37.5% (160/426) in 2019, 28.2% (120/426) in 2020 and 37.27% (146/426) in 2021. These results coincide to those of Chih-Cheng and al in patients of Veterans General Hospital of Taiwan who had also found a decreasing number of
Streptococcus pneumonia, Staphylococcus aureus, Enterococcus spp, Escherichia coli and
Klebsiella pneumonia from 2019 to 2020 [
7]. The high number of strains isolated in the pre-CoVID19 period (2019) constrasting with the lowest amount in the year of COVID onset (in 2020) is logical and could rely on the wide and systematic implementation of COVID-19 barriers measures including hygienic measures, wearing of face mask, social distancing that lead to rapid and overall decline of respiratory tract infections as well as feco-oral transmitted diseases [
8] In fact, giving similar routes of transmission, all barriers measures applicable for COVID-19 have the advantage to also block others infections [
9] Furthermore, the occurrence of the COVID-19 pandemic led to an overconsumption of antibiotics as well as self-medication, which in turn destroyed the lot of potential pathogenic and commensal bacterial populations [
8].
Concerning the bacterial profile, 51.4% (219/426) of the included bacteria isolates were
Enterobacteriaceae, 45% (192/426)
Staphylococcus strains, 3% (13/426)
Streptococcus strains and 0.3% (2/426) of
Neisseria meningitides with relatively high rate of resistance over time. Chih-Cheng also found
Enterobacteriaceae as the most isolated bacteria 30.8% (49/159) in patients at Veterans General Hospital of Taiwan between 2019 and 2020[
8]. Others previous studies worldwide have also reported the predominance of
Enterobacteriaceae in clinical samples [
11,
13,
15]. In fact,
Enterobacteriaceae are the most incriminated commensal germs in opportunistic infections in general [
11,
15]. In this study, the main isolated species among
Enterobacteriaceae were
Klebsiella pneumoniae 41.1% (90/219),
E. coli 35.6% (78/219),
Enterobacter 11.4% (25/219) and
proteus mirabilis 9.13% (20/219), similar trends has been already observed in previous studies [
11,
15]. In fact, the rapid emergence of bacteria especially resistant bacteria is occurring worldwide, endangering the efficacy of antibiotics, which have transformed medicine and saved millions of lives [
16,
17,
18,
19]
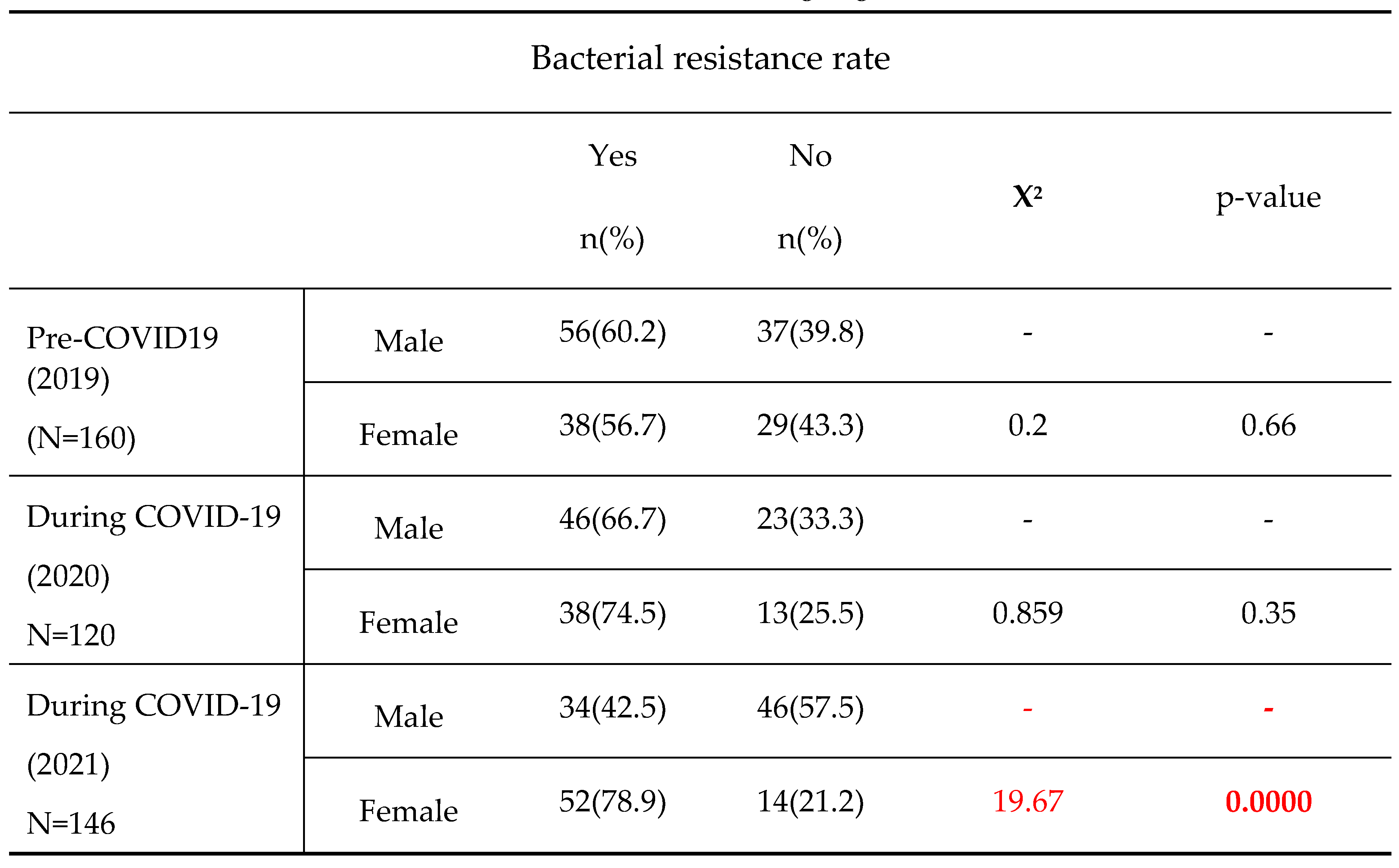
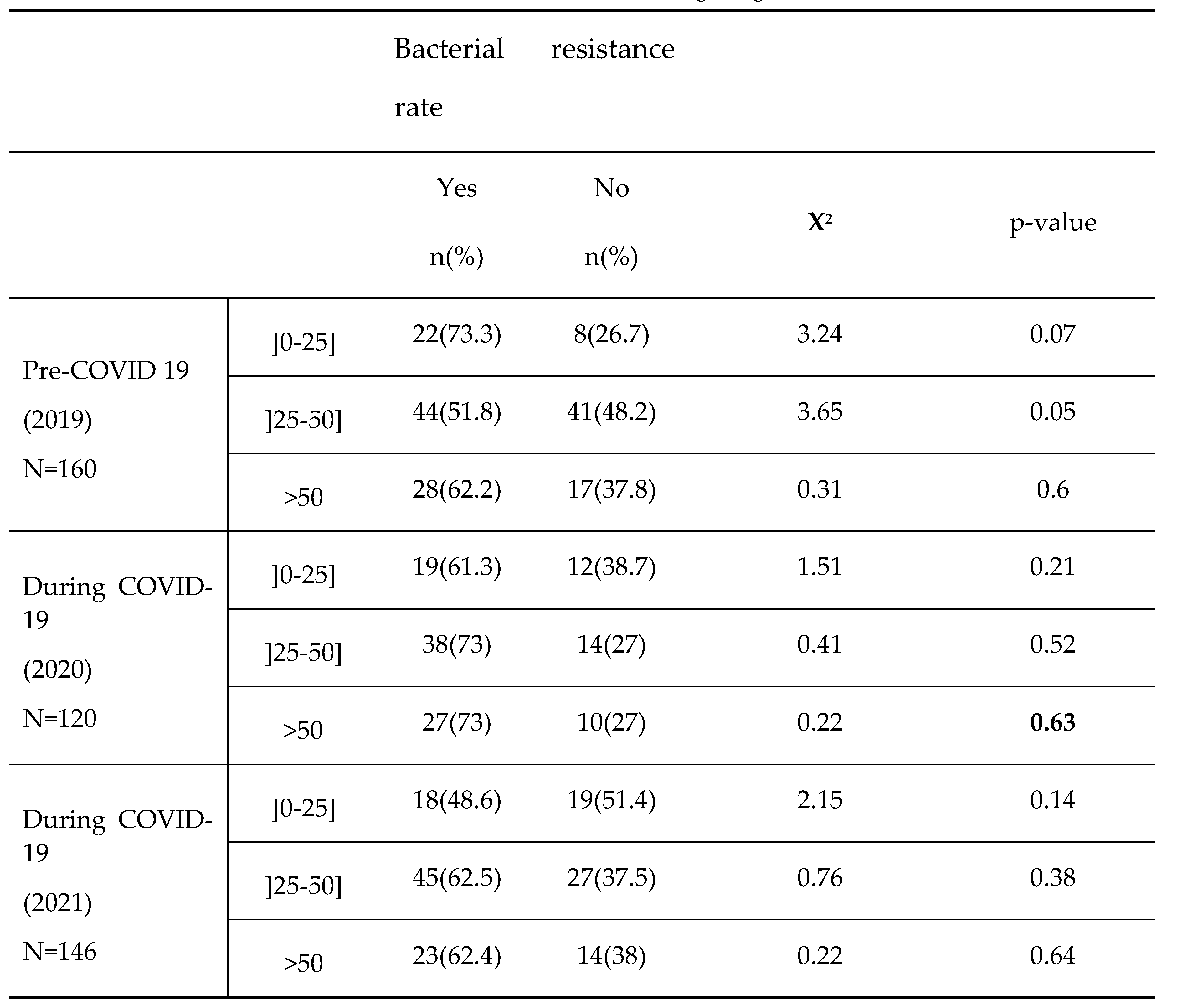
In this study, bacterial resistance tends to increase in female’s subjects over years. Moreover, In comparison to subjects aged ]0-25] with decreasing resistance rate over time, the subjects aged ]25-50] and >50 more likely exhibited relatively higher resistance with the peak of resistance recorded in 2020, the year of COVID-19 onset, even though the observed differences were not statistically significant. Previous studies already reported a high prevalence of bacterial resistant strain either with ages [
14] and or in female’s subjects [
1]. This disproportionate burden of antimicrobial resistance (AMR) on women is due to both demand and supply-side factors. Some demand-side factors which increase women’s vulnerability to AMR are biological factors, women’s nature and type of employment, excessive home-based care work, and limited access to healthcare [
20](WHO, 2018). On the supply-side, gender differences in antibiotic prescription by doctors due to lack of training and gender-bias increase women’s antibiotic usage (AMU) [
21,
22]. In fact, Women are 27% more likely to receive an antibiotic prescription in their lifetime compared to men [
23]. Furthermore, the high rate of antimicrobial resistance in subject aged >25 years during the COVID-19 pandemic in this study is not surprising. This aged range corresponds to the most active part of the Cameroonian population who is believed to have taken the highest amount of medications including antibiotics to protect against COVID-19 at work as the pandemic evolved in 2020.
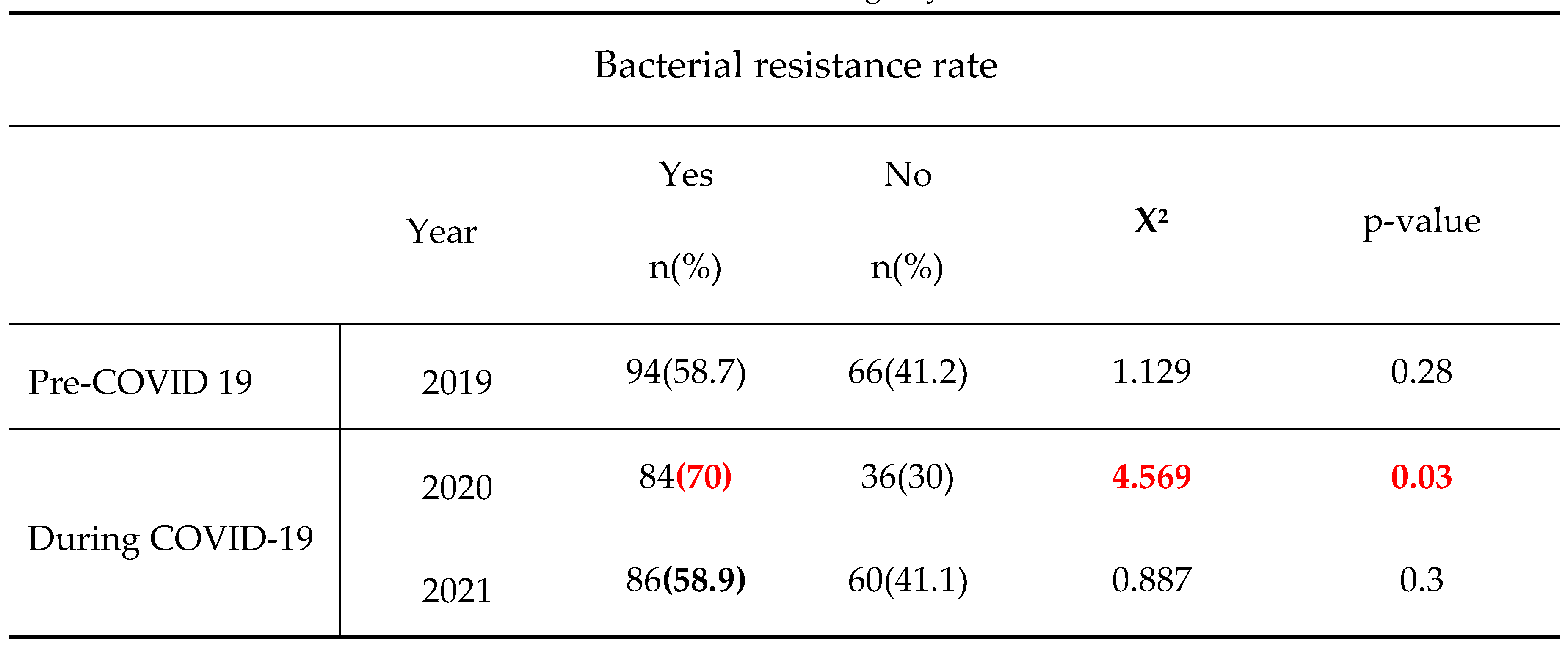
This study also showed that in 2019, corresponding to the pre-COVID-19 period, the highest number of bacteria isolates and lowest rate of bacterial resitance were recorded (160 isolates vs. 58.8% resistance rate). However, we noticed in contrast lower bacteria isolated during COVID-19 pandemic era but greater resistance burden with the lowest bacteria amount and peak of bacteria resistance registered in 2020, the year of COVID-19 onset (120 isolates vs. 70% resistance in 2020 and 146 isolates vs.58.9% resistance in 2021). Multiple studies reported an unexpected high incidence of infections due to methicillin-resistant
S. aureus, carbapenem-resistant
A. baumannii, carbapenem-resistant
Enterobacteriaceae among COVID-19 patients admitted to the intensive care unit [
9,
15]. Moreover, a metanalysis Of 1331 articles of Ruwandi shows that during the first 18 months of the pandemic, AMR prevalence was high in COVID-19 patients and varied by hospital and geography although there was substantial heterogeneity [
10]. According to ruwandi et al, the increase in the prevalence of resistant strains in 2020 could be straightly related to the peak of antibiotic consumption during the COVID-19 pandemic onset, ranging up to 74.7 % from 2019 to 2020 for antibiotics such as Fosfomycin [
10]; that generally increased the selection pressure and evolvement of resistance mechanism [
10].
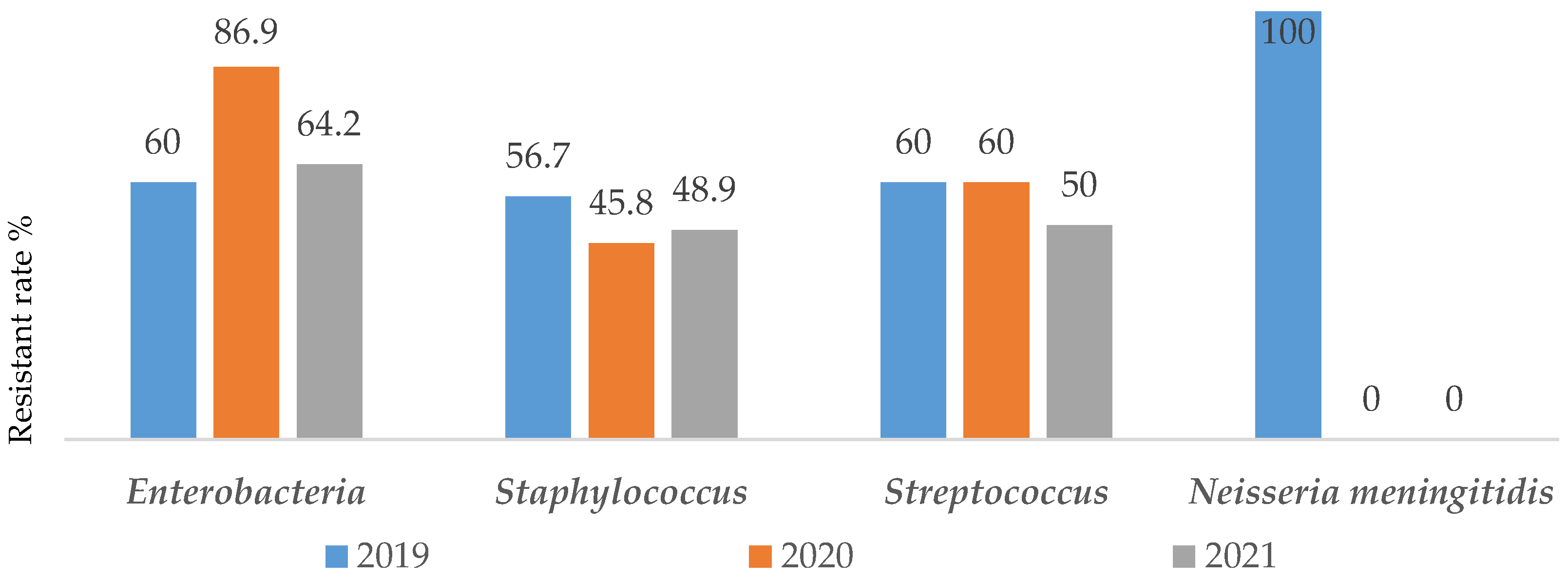
In constrat to almost all others groups of bacteria where the resistance burden was quite constant over years,
Enterobacteriaceae exhibited greater resistance rate during the pandemic period [60%(48/80) in 2019 vs 86.9%(60/69) in 2020 and 64.2%(61/95) in 2021)]. Furthermore, no resitance to antibiotics has been noted concerning
Neisseria meningetidis in 2020 and 2021 [100%(1/1) in 2019 vs 0% of strain in 2020 and 2021)]. A similar study on the impact of SARS-CoV-2 epidemic on antimicrobial resistance shows that, the Gram-negative bacteria were isolated from 78% of patients with predominant
Enterobactericaea resistant strains including carbapenemases producers such as K. pneumoniae (92.6%) and A. baumannii (72.8%)[
11]. This result is not different from that of Sanofi, which estimates that 30% of Klebsiella pneumoniae strains show resistance to 3rd generation cephalosporins in France, in 2020 [
12]. Unlike
Neisseria meningetidis, Enterobacteriaceae are widely distributed and have a large host range [
24], they can cross-infect and spread between medical staff and patients, and also their genetic materials (such as plasmids or transposons) can be obtained from the outside world, leading to horizontal transmission of drug-resistant genes, which further leads to the wide spread of drug-resistant bacteria [
25,
26].
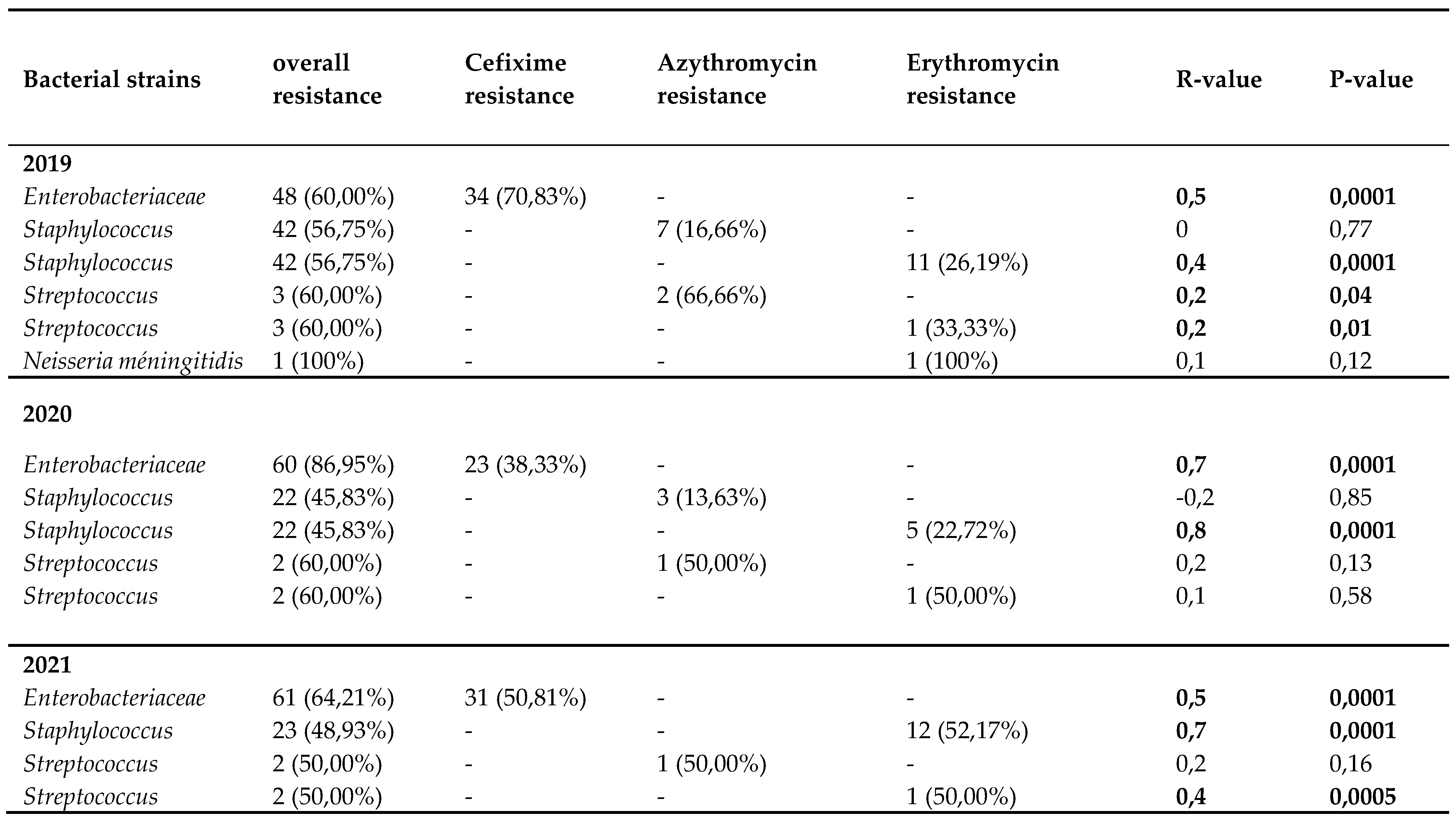
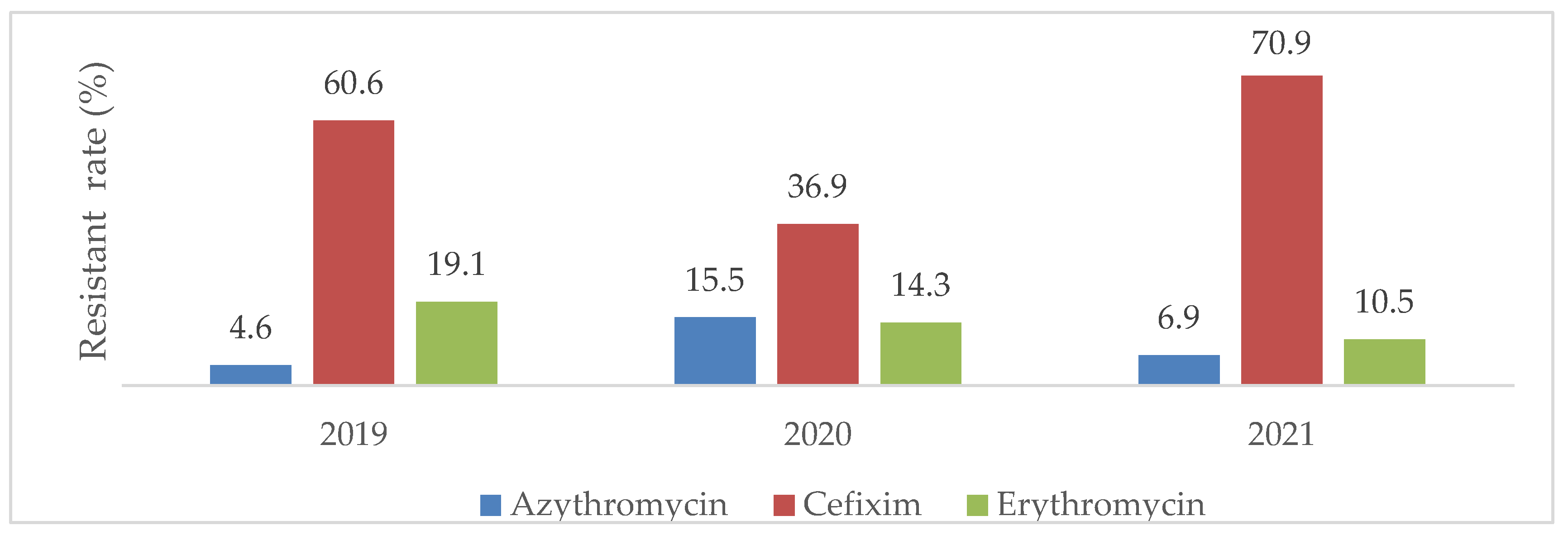
The resistance of each bacteria isolate to its selected antibiotic tends to slightly decrease the year of the COVI-19 pandemic onset (2020) before re-increasing or staying constant one year thereafter (2021). A significant association was found between resistant
Enterobacteriaceae strains and Cefixime (R= 0.7; P-value= 0.0001) and also, between resistant
Staphylococcus strains and Erythromycin (R= 0.8; P-value= 0.0001). Chih-Cheng et al reported in 2020 Taiwan a rapid increase in multidrug-resistant organisms (MDROs), including extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae, carbapenem-resistant New Delhi metallo-β-lactamase (NDM)-producing Enterobacterales, Acinetobacter baumannii, methicillin-resistant Staphylococcus aureus (MRSA) [
7]. Besides, several recent reports have described an increase in multidrug-resistant organisms (MDROs) during the COVID-19 pandemic [
27,
28,
29,
30]. The cause of this high resistance is multifactorial and is particularly related to high rates of antimicrobial agent utilization in COVID-19 patients with a relatively low rate of co- or secondary infections [
31]. This clearly confirms that anticromicrobial resistance should be continuously closely-monitor during and after COVID pandemic era.
4. Materials and Methods
4.1. Study duration and location
A retrospective cross-sectional and analytical study was conducted at the Central Hospital and the General Hospital of Yaounde from September 2021 to July 2022. Data from registries were collected over a period of 3years from 1 January 2019 to 31 December 2021.
4.2. Sampling method and Sampling population
Our study focused on bacteria Streptococcus, Staphylococcus, Haemophilus influenzae, Neisseria meningitidis and Enterobacteriaceae resistant or not to Cefixime, Azythromycin and Erythromycin; isolated from samples of patients consulting the bacteriology laboratories of the Central and General hospitals of Yaoundé from January 2019 to December 2021, archived in the annual registers where the data is organized by week and month with all the information on the pre-analytical information, analytical and post-analytical. We carried out a census on the basis of already existing data. All the bacteria Streptococcus, Staphylococcus, Haemophilus influenzae, Neisseria meningitidis and Enterobacteriaceae resistant to Cefixime and to Azythromycin and Erythromycin recorded in the results registers of the bacteriology department were sampled. Indeed, the recording of the results in the registers that we questioned was done according to a fairly rigorous protocol, by providing information on the patient sampled, the type of sample taken, the dates of sampling and analysis, the techniques analysis used, the germs identified, the results of the antibiogram for each germ isolated. We therefore extracted the data day after day by questioning the registers to obtain the information that should allow us to meet our objectives.
4.3. Statistical analysis
Data were collected using Kobotolbox software, entered into a Microsoft Excel 2016 database (version 15.13.3) and analyzed using StatView software (version 5.0.0.0) and the simple linear regression statistical test a was used to show the relationships between the antibiotics of interest tested and the resistant bacterial strains isolated and the Chi-Square of independence to compare the carriage prevalences of resistant bacterial strains from 2019 and 2021 to that of the year of the onset of the pandemic to COVID19.
Data such as age, sex, location of patients whose samples were subjected to bacteriological analyzes were presented as a percentage. Also the distribution of the frequency of isolated bacteria, the genus to which they belong, the antibiotics to which the bacteria were resistant were presented in strip diagram and tables. The resistance profile of the strains as well as their relationship with the antibiotics of interest were presented in tables and the resistant strain and antibiotic relationships were established via simple linear regression. We have made an annual presentation of the overall prevalence of carriage of selected resistant bacteria, spread over each of the 03 years, on a vertical band and curve diagram. Statistical comparisons of the variation in the prevalence of the carriage of resistant bacterial strains in 2019 and 2021 with that of the year of the onset of the COVID19 pandemic (2020), were made by the Chi-Square test and presented in tables.
Author Contributions
Conceptualization, Cecile Ingrid Djuikoue, Willy Yamdeu Djonkouh, Rodrigue Kamga Wouambo and Benjamin D. Pokam; Data curation, Cecile Ingrid Djuikoue, Willy Yamdeu Djonkouh, Cavin Epie Bekolo, Paule Dana Djouela Djoulako and Gilder Tonfack Temgoua; Formal analysis, Willy Yamdeu Djonkouh, Cavin Epie Bekolo, Rodrigue Kamga Wouambo and Paule Dana Djouela Djoulako; Investigation, Willy Yamdeu Djonkouh, Rodrigue Kamga Wouambo and Gilder Tonfack Temgoua; Methodology, Cecile Ingrid Djuikoue, Paule Dana Djouela Djoulako and Benjamin D. Pokam; Project administration, Cecile Ingrid Djuikoue, Cavin Epie Bekolo, Benjamin D. Pokam, Nicolas Antoine-Moussiaux and Teke R. Apalata; Resources, Raspail Carrel Founou, Gilder Tonfack Temgoua, Nicolas Antoine-Moussiaux and Teke R. Apalata; Software, Willy Yamdeu Djonkouh, Paule Dana Djouela Djoulako and Gilder Tonfack Temgoua; Supervision, Raspail Carrel Founou and Teke R. Apalata; Validation, Paule Dana Djouela Djoulako and Teke R. Apalata; Visualization, Cecile Ingrid Djuikoue, Willy Yamdeu Djonkouh, Rodrigue Kamga Wouambo, Raspail Carrel Founou and Benjamin D. Pokam; Writing – original draft, Cecile Ingrid Djuikoue, Willy Yamdeu Djonkouh, Rodrigue Kamga Wouambo and Paule Dana Djouela Djoulako; Writing – review & editing, Cecile Ingrid Djuikoue, Willy Yamdeu Djonkouh, Cavin Epie Bekolo, Rodrigue Kamga Wouambo, Raspail Carrel Founou, Paule Dana Djouela Djoulako, Gilder Tonfack Temgoua, Benjamin D. Pokam, Nicolas Antoine-Moussiaux and Teke R. Apalata.