1. Summary
The sclera of the eye and body skin discoloration to yellow are the main apparent symptoms of neonatal jaundice [
1]. Jaundice is caused due to high levels of bilirubin in the patient's blood due to premature liver, known as hyperbilirubinemia [
2]. The severity of hyperbilirubinemia classifies it as one of the top causes of newborns' mortality and permanent disorders causes. As statistics implied, over a decade ago, hyperbilirubinemia caused 114000 deaths and 75000 brain dysfunction in newborns [
3]. Diagnosis of hyperbilirubinemia can be achieved by collecting blood samples from the patient, and this specific test is called Total Serum Bilirubin (TSB) [
4]. This type of test causes stress and discomfort to the patient since it is an invasive method; hence, a non-invasive way is preferred. Transcutaneous Bilirubin (TcB) is a non-invasive technique for bilirubin calculation [
5]; however, this method is not available in all healthcare institutes [
6].
Researchers started implementing image-processing techniques for diagnosing jaundice more than two decades ago. In 2009, a study by Leartveravat [
7] attempted to calculate bilirubin levels for 61 neonates with jaundice non-invasively by analyzing their images taken by a digital camera using the CMYK calculation method. The researcher calculated the CMYK components manually using photoshop and estimated the bilirubin levels by subtracting values of M from Y components. After implementing Pearson’s product-moment and linear regression methods, they discovered a significant correlation between bilirubin levels measured using TSB and the Y-M value. This method was the start of many attempts to diagnose jaundice using non-invasive techniques despite being an approximation method lacking precision. The researchers Mansour et al. [
8] attempted to diagnose hyperbilirubinemia or jaundice using color detection. They used images from a random database of infant monitoring from the google website. They picked the pictures from the image acquisition toolbox in Matlab of normal and jaundiced infants with different lighting environments and capturing angles. They used the YCrCb color space method to exclude the Y component, which represents the luminance, and the CrCb component, which represents the chrominance due to being stored in different channels. Then, standard deviation, mean, and kurtosis methods were used to compare the skin colors of normal and jaundiced infants. Munkholm et al. [
9] proposed TcB measurement from images taken with a dermatoscopy attached to iPhone 6 camera with a Wratten No. 11 filter placed in between. Pearson’s correlation coefficient was used to evaluate the relation between intensity and TSB levels. Regardless of their results, they were able to collect a total of 64 infant images only. Another study by Endang et al. [
10] proposed a system used in estimating the risk zone for jaundiced neonates based on skin color analysis. They used a digital camera for capturing newborn images. They collected 120 images only and performed multiple techniques to acquire values of RGB, HSV, and YCbCr color spaces and used them as input parameters for modeling and validation of linear regression. They achieved 67% accuracy. On the other hand, Padidar et al. [
11] proposed a method for jaundice detection using a mobile application for Android. The results were promising, although they managed to collect only 113 infant images. Ayden et al. [
12] used AI as a classifier was made in 2016. They used only 80 images of infants; half were for normal infants, and the other half were for jaundiced ones, all taken by a smartphone camera. They used an image segmentation technique to achieve color balance using an 8-colored card put on a specific area of the baby’s skin for calibration. Afterward, they used color map transformation and feature extraction on the baby’s skin color and the calibration card in RGB, YCrCb, and LAB color space. Then, kNN (k-Nearest Neighbour) and SVR (Support Vector Regression) algorithms were applied to the acquired data to estimate the bilirubin levels. These results show better quality results with less processing time using AI techniques.Recently, Hashim et al. [
13] attempted to use image-processing methods for diagnosing jaundice. They used only two manikins and 20 images of infants due to the unavailability of more neonate images.
All previous works mentioned used infant images of no more than 120 due to the difficulty and scarceness of obtaining such images. This data article provides 670 neonate images, which acts as a valuable source of data for developing studies in jaundice detection and AI techniques to assist medical staff in the NICU in diagnosing jaundice accurately and non-invasively quickly.
2. Data Description
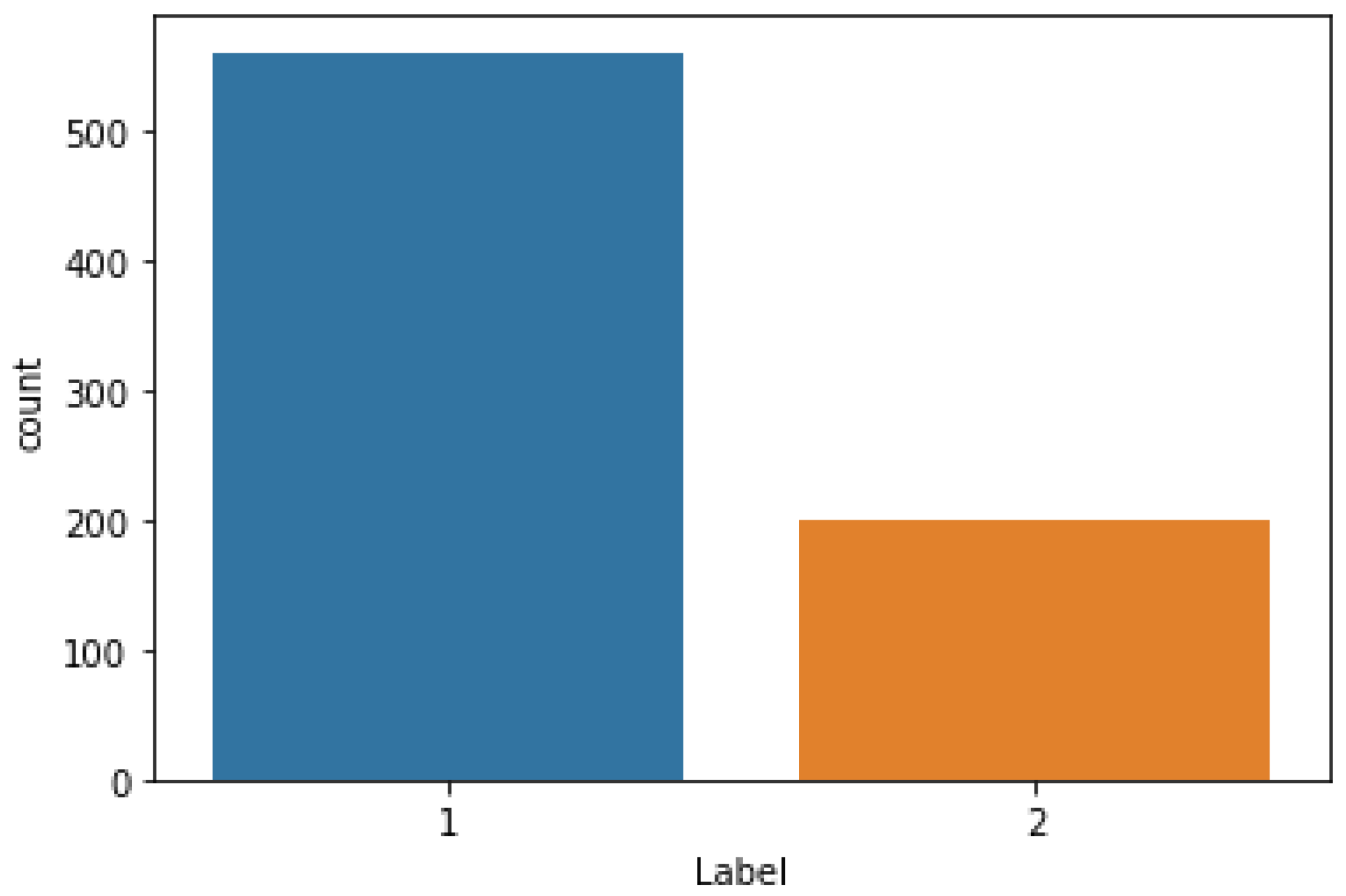
This dataset article provides images of newborns taken in the NICU at Al-Elwiya Maternity Teaching Hospital in Al Rusafa, Baghdad, Iraq. It is a hospital specializing in obstetrics and gynecology; therefore, all infants are considered aseptic. This data comprises normal and jaundiced infant images from different angles and lighting environments. Thus, collecting as many images as possible helps increase the accuracy. The collected data includes 670 infant images (560 normal and 200 jaundiced) with 1000 × 1000 resolution, all in jpg format. The images were taken by an iPhone 11 pro max 12 MP camera. The dataset is composed of three folders: normal neonate images, jaundiced neonate images, and an excel sheet file in CSV (Comma delimited) format that contains the RGB and YCrCb channel values in addition to the status of each row values, either “1” for normal, or “2” for jaundiced. The classification of NJN data and specification table are shown in
Figure 1 and
Table 1, respectively.
3. Methods
2.1. Ethics Considerations
The data was collected from about 600 newborns aged between 2 to 6 days with different skin tones and weights. All infant image data were collected from Al-Elwiya Maternity Teaching Hospital in Al Rusafa, Baghdad, Iraq, all according to the Declaration of Helsinki guidelines (Finland 1964) with the clearance ethics granted by the research committee in Al Rusafa Directorate of Health, Iraqi Ministry of Health and Environment, Baghdad, Iraq (Protocol number: 2022019) and written approval of the legal guardian for each infant.
2.2. Data Evaluation
The experimental assessment was carried out using the Python program (version 3.9) in the Spyder integrated development environment (IDE) (version 5.2.2) from the Anaconda3-Navigator. To evaluate the collected data, the color intensity values of RGB and YCbCr obtained from the selected ROI from each infant have been collected and placed on an excel file (train.csv). The evaluation metrics, including accuracy, precision, recall, F1-score, and confusion matrix, were used for evaluating the data based on three AI techniques, including K-Nearest Neighbors (KNN)[
13], Random Forest (RF) [
14] and eXtreme Gradient Boosting (XGboost) [
15]. All these techniques used 80% of the data for training and 20% for testing and provided the weighted average of the above metrics, as shown in
Table 2.
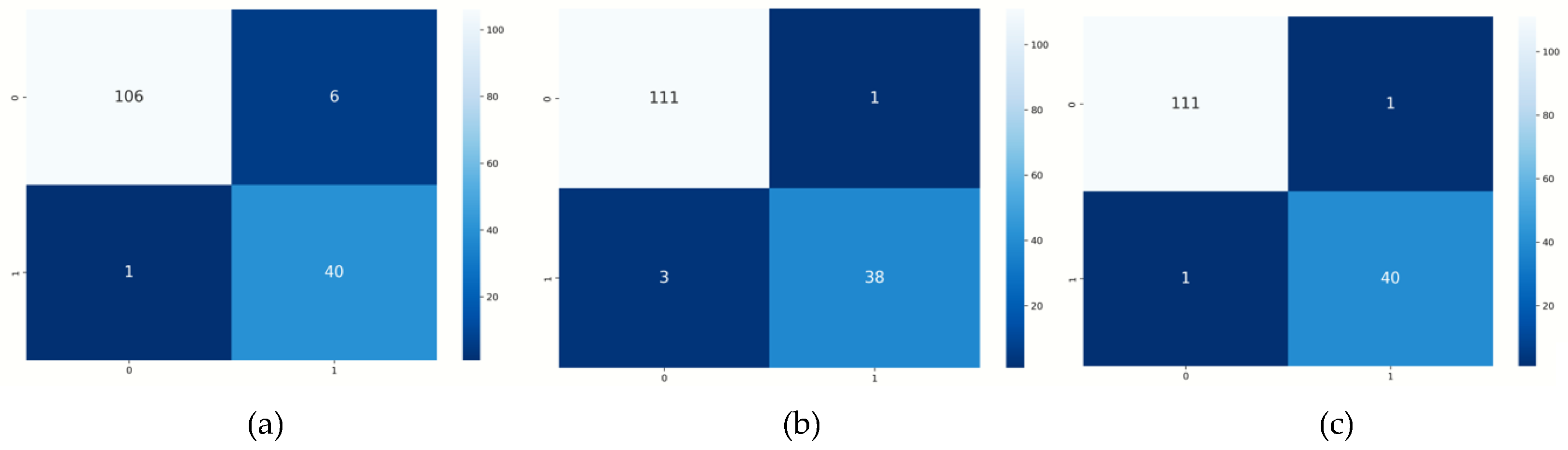
From
Table 2, we have observed that XGboot has the best accuracy amongst all the techniques used in this work, with an accuracy of 98.6%, and KNN having the least accuracy of 95.4%.
The visualization of the confusion matrix from three AI techniques is shown in
Figure 2 having the number of instances of True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN).
4. User Notes
Images of normal and jaundiced neonates are scarce online and not easily accessible.
Healthcare professional developers working in the AI field can benefit from this data.
Other researchers in biomedical engineering and computer science can also use the provided images in skin color analysis for neonates to diagnose jaundice or other skin conditions.
Provided images comprise 560 normal and 200 jaundiced infants.
The images are in jpg format with 1000 × 1000 resolution.
An excel sheet in CSV (comma delimited) format that contains RGB and YCbCr channel values for all the provided images.
Author Contributions
Conceptualization, A.A-N.; methodology, A.Y.A, A.A-N and S.L.M; software, A.Y.A, and A.A-N; validation, A.Y.A, and A.A-N; investigation, A.Y.A, A.A-N and S.L.M; resources, A.Y.A.; data curation, A.Y.A.; writing—original draft preparation, A.Y.A, and A.A-N.; writing—review and editing, A.Y.A, A.A-N and S.L.M.; visualization, A.A-N.; supervision, A.A-N and S.L.M.; project administration, A.A-N and S.L.M.; funding acquisition, A.A-N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the research committee in Al Rusafa Directorate of Health, Iraqi Ministry of Health and Environment, Baghdad, Iraq (Protocol number: 2022019) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Acknowledgments
The authors show their gratitude and appreciation to Middle Technical University, Electrical Engineering Technical College-Baghdad, Iraq, for the support and encouragement for disseminating scientific engineering research and to Al Elwiya Maternity Teaching Hospital for providing the required data to perform this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dzulkifli, F.A.; Mashor, M.Y.; Khalid, K. Methods for determining bilirubin level in neonatal jaundice screening and monitoring: A literature review. Journal of Engineering Research and Education 2018, 10, 1-10.
- Brits, H.; Adendorff, J.; Huisamen, D.; Beukes, D.; Botha, K.; Herbst, H.; Joubert, G. The prevalence of neonatal jaundice and risk factors in healthy term neonates at National District Hospital in Bloemfontein. African Journal of Primary Health Care and Family Medicine 2018, 10, 1-6. [CrossRef]
- Bhutani, V.; Zipursky, A.; Blencowe, H.; Khanna, R.; Sgro, M.; Ebbesen, F.; Bell, J.; Mori, R.; Slusher, T.; Fahmy, N. du L, Okolo AA, de Almeida MF, Olusanya BO, Kumar P, Cousens S, Lawn JE (2013) Neonatal hyperbilirubinemia and rhesus disease of the newborn: incidence and impairment estimates for 2010 at regional and global levels. Pediatr Res 2013, 74, 86-100. [CrossRef]
- Mishra, S.; Agarwal, R.; Deorari, A.K.; Paul, V.K. Jaundice in the newborns. The Indian Journal of Pediatrics 2008, 75, 157-163. [CrossRef]
- Pediatrics, A. Management of hyperbilirubinemia in the newborn infant 35 or more weeks of gestation. Am. Acad. Pediatr 2004, 114, 297-316. [CrossRef]
- Mantagou, L.; Fouzas, S.; Skylogianni, E.; Giannakopoulos, I.; Karatza, A.; Varvarigou, A. Trends of transcutaneous bilirubin in neonates who develop significant hyperbilirubinemia. Pediatrics 2012, 130, e898-e904. [CrossRef]
- Leartveravat, S. Transcutaneous bilirubin measurement in full term neonate by digital camera. Medical Journal of Srisaket Surin Buriram Hospitals 2009, 24, 105-118.
- Mansor, M.; Yaacob, S.; Hariharan, M.; Basah, S.; Jamil, S.A.; Khidir, M.M.; Rejab, M.; Ibrahim, K.K.; Jamil, A.A.; Junoh, A. Jaundice in newborn monitoring using color detection method. Procedia Engineering 2012, 29, 1631-1635. [CrossRef]
- Munkholm, S.B.; Krøgholt, T.; Ebbesen, F.; Szecsi, P.B.; Kristensen, S.R. The smartphone camera as a potential method for transcutaneous bilirubin measurement. PloS one 2018, 13, e0197938. [CrossRef]
- Juliastuti, E.; Nadhira, V.; Satwika, Y.W.; Aziz, N.A.; Zahra, N. Risk zone estimation of newborn jaundice based on skin color image analysis. In Proceedings of the 2019 6th International Conference on Instrumentation, Control, and Automation (ICA), 2019; pp. 176-181. [CrossRef]
- Padidar, P.; Shaker, M.; Amoozgar, H.; Khorraminejad-Shirazi, M.; Hemmati, F.; Najib, K.S.; Pourarian, S. Detection of neonatal jaundice by using an android OS-based smartphone application. Iranian Journal of Pediatrics 2019, 29. [CrossRef]
- Hashim, W.; Al-Naji, A.; Al-Rayahi, I.A.; Oudah, M. Computer vision for jaundice detection in neonates using graphic user interface. In Proceedings of the IOP Conference Series: Materials Science and Engineering, 2021; p. 012076. [CrossRef]
- Kramer, O.; Kramer, O. K-nearest neighbors. Dimensionality reduction with unsupervised nearest neighbors 2013, 13-23.
- Fawagreh, K.; Gaber, M.M.; Elyan, E. Random forests: from early developments to recent advancements. Systems Science & Control Engineering: An Open Access Journal 2014, 2, 602-609. [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T. Xgboost: extreme gradient boosting. R package version 0.4-2 2015, 1, 1-4.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).