2.2. Data
The choice of data is dependent on its intended usage. For our research aimed at recognizing agricultural land cover types, we needed to consider the phenological stage of the crops on the agricultural plots. Based on a promising suggestion in the literature, we decided to investigate the feasibility of using data collected approximately four weeks before harvest. In Poland, the harvest season typically lasts for 2 months (July and August), starting with the small harvest of rapeseed and winter barley, followed by the large harvest of rye, spring barley, wheat, and oats [
25]. To examine the potential of using a single registration for agricultural land cover recognition, a survey campaign was conducted in 2021 that collected the following data: S2 time series, one-shot hyperspectral, in-situ measurement, and topographical data. The data acquisition dates are as follows:
Sentinel-2 time series during vegetation season 2021 (March-September)
Hyperspectral data: 5th of July 2021
in-situ measurements: 7 July 2021
Shuttle Radar Topography Mission - SRTM
archival topographic data existing in Central Geodetic and Cartographic Resource
During the 2021 growing season, only six Sentinel-2 registration dates were cloud-free:
S2B_MSIL2A_20210327T093039_N0214_R136_T34UEA_20210327T120034
S2A_MSIL2A_20210411T093031_N0300_R136_T34UEA_20210411T122810
S2B_MSIL2A_20210509T094029_N0300_R036_T34UEA_20210509T120133
S2B_MSIL2A_20210725T093039_N0301_R136_T34UEA_20210725T115620
S2B_MSIL2A_20210728T094029_N0301_R036_T34UEA_20210728T125908
S2B_MSIL2A_20210906T094029_N0301_R036_T34UEA_20210906T113414
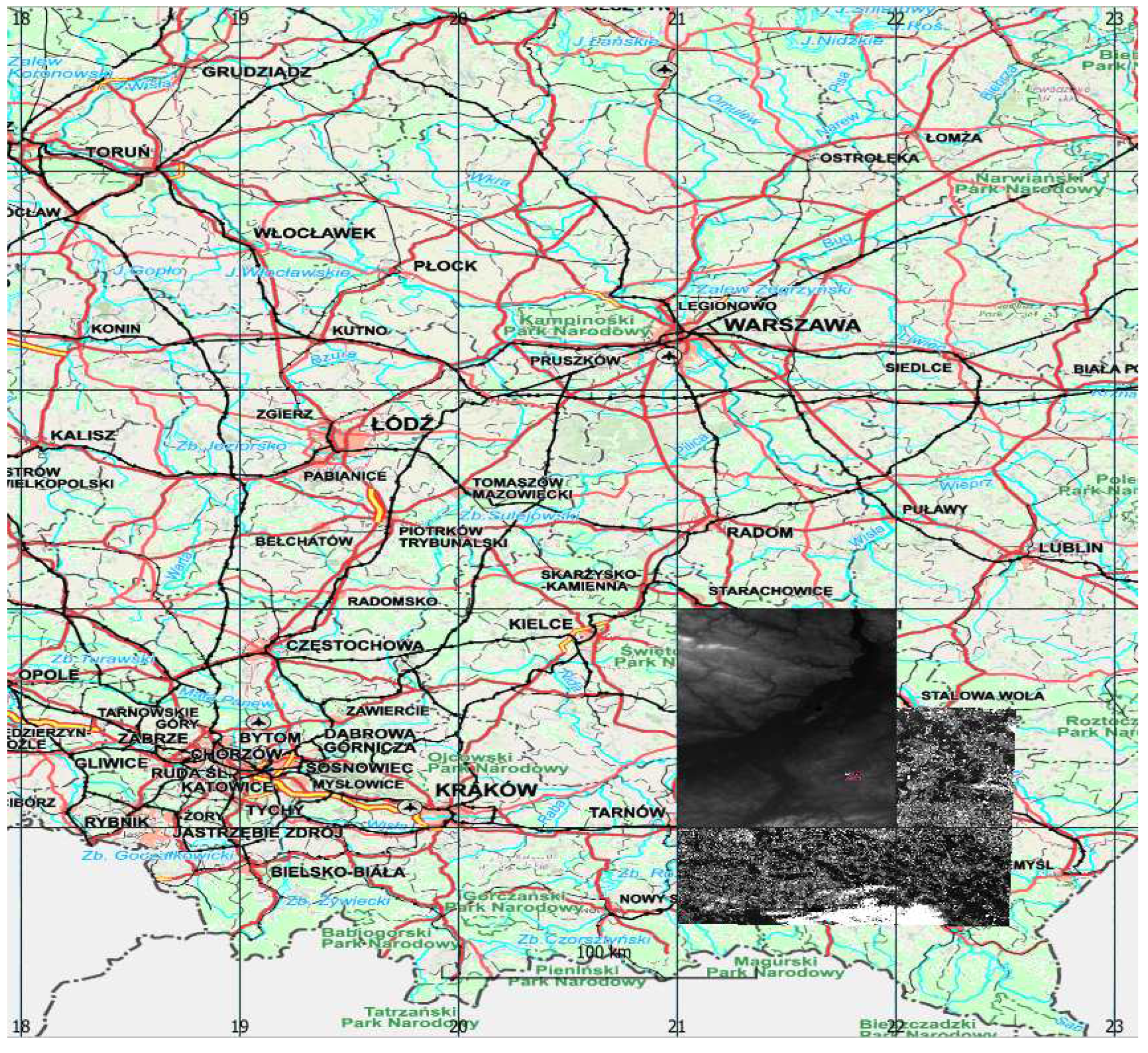
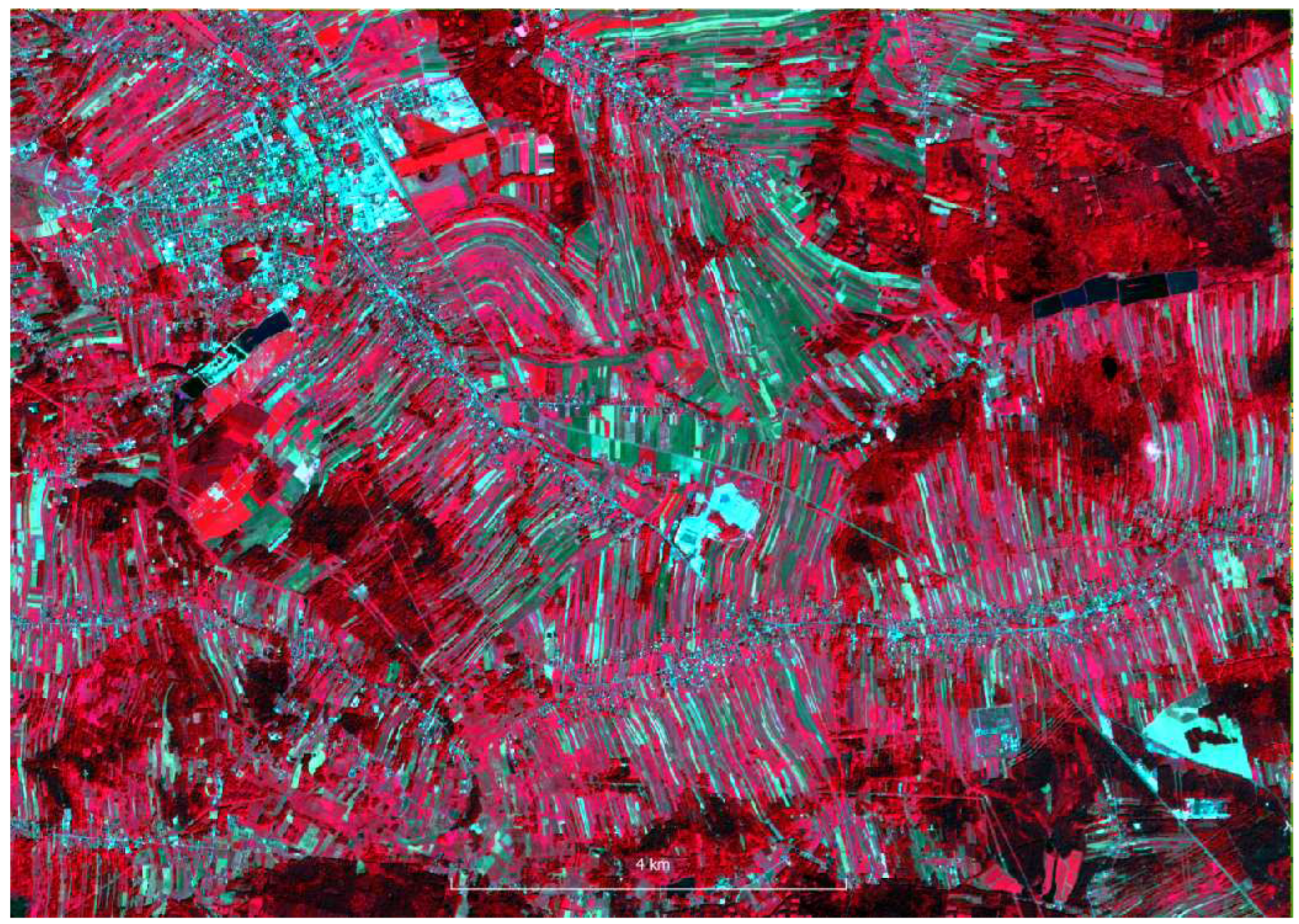
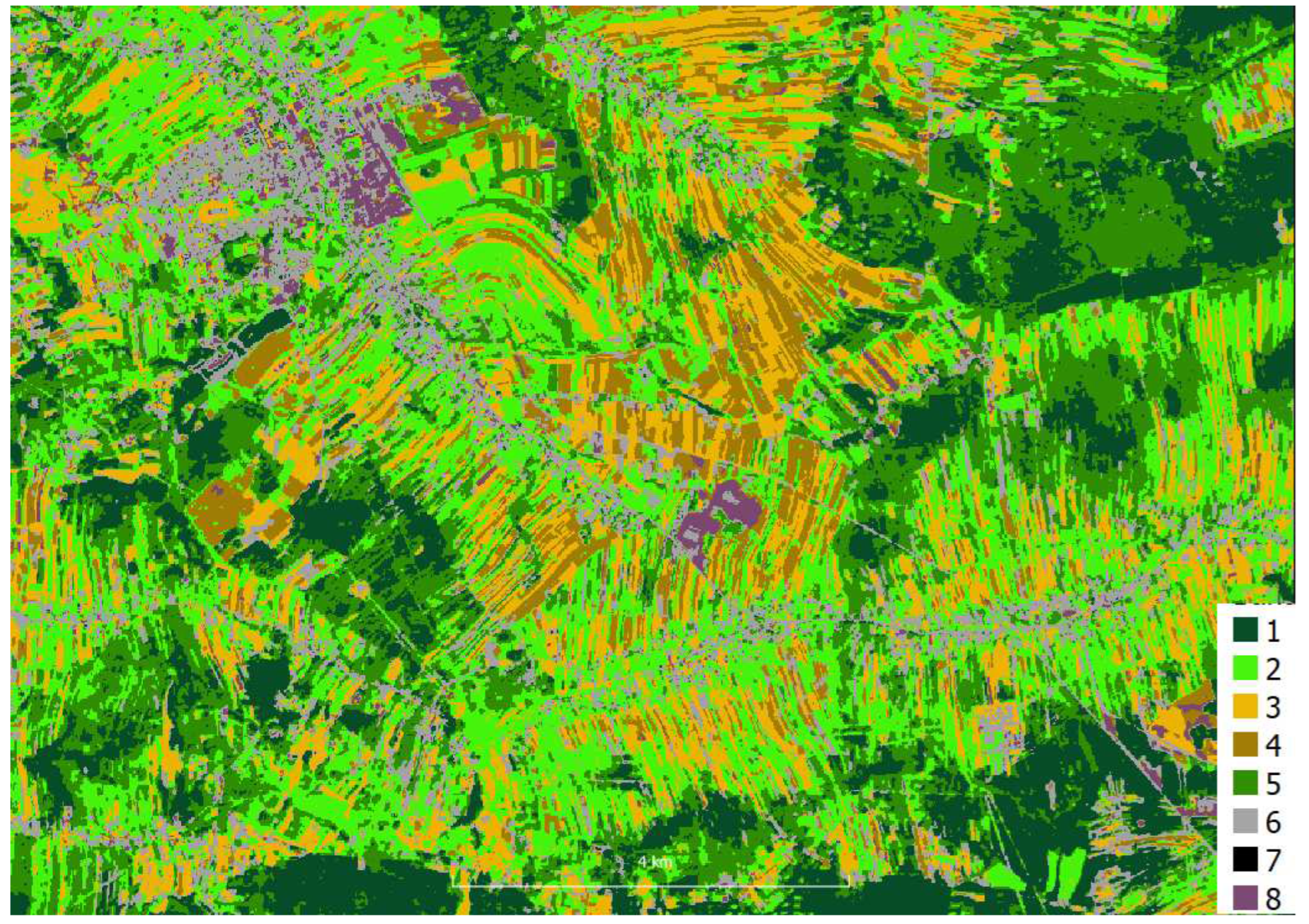
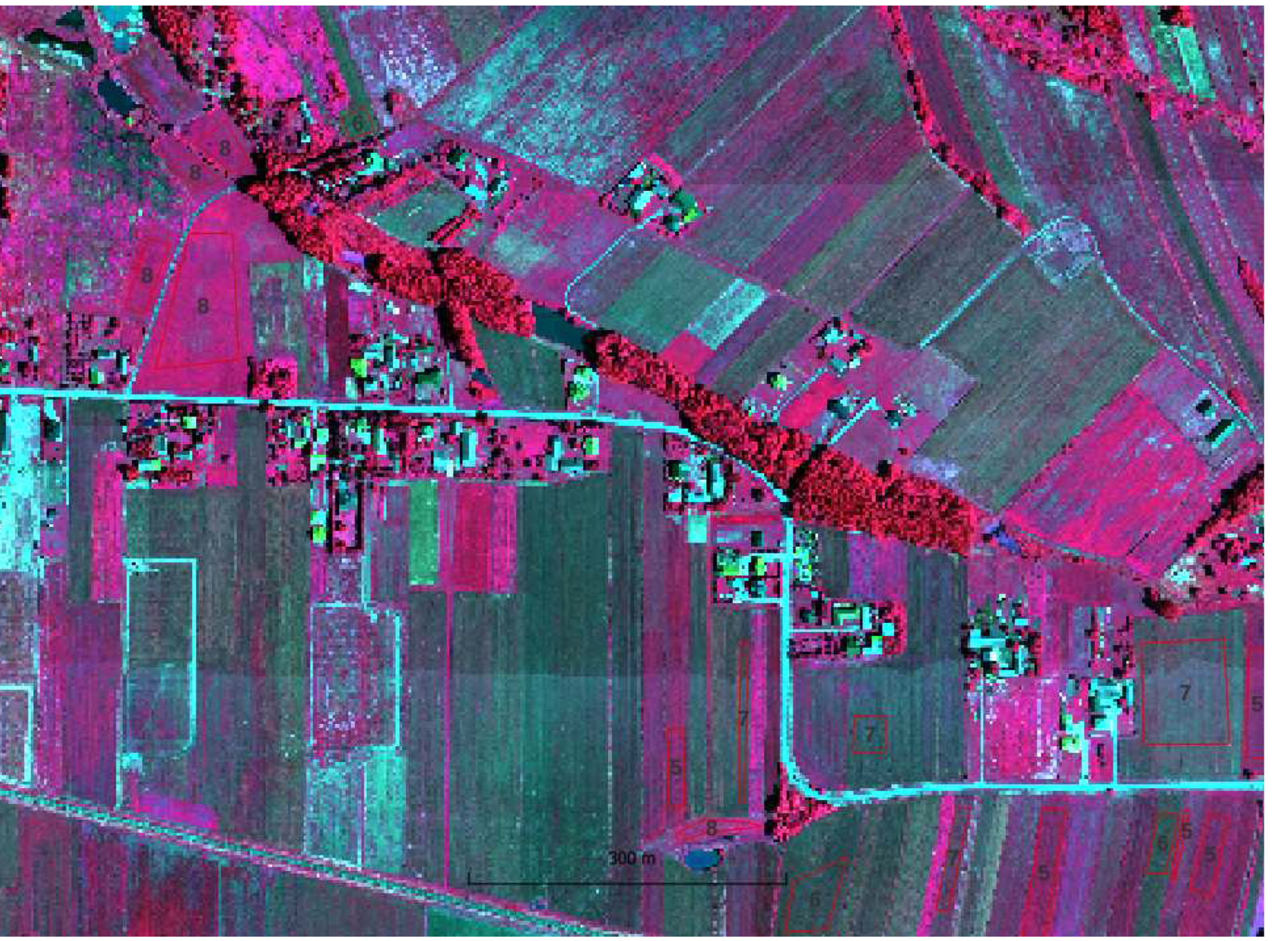
Sentinel-2 images were acquired from Copernicus Open Access Hub as granules with a size of 100 per 100 km with a radiometric correction level of 2A in geographical coordinate system EPSG:4326. The images were not further corrected either geometric or radiometric. The pixel size depending on the channel is 10, 20 and 60m. A single S2 scene in SAFE (ESA) format takes approximately 1.2 gigabytes when packed (S2 range in shown in
Figure 1).
Hyperspectral data were acquired for the area ca. 5 x 4 km using HySpex VS-725 which is a very small area compared to the S2 range (in red in
Figure 1) . The registration was performed at an altitude of 867 - 882 m. The HySpex VS-725 consists of two SWIR-384 scanners and one VNIR-1800 scanner which provide 430 spectral channels (414.13 nm - 2357.43 nm). The test area was covered with 16 strips. Radiometric, geometric (PARGE), atmospheric (ATCOR4) correction was performed using the MODTRAN physical model. The final product, an orthophotomap with a pixel size of 0.5 m was registered in the UTM 34N coordinate system (EPSG:32634) and takes up about 60 gigabytes.
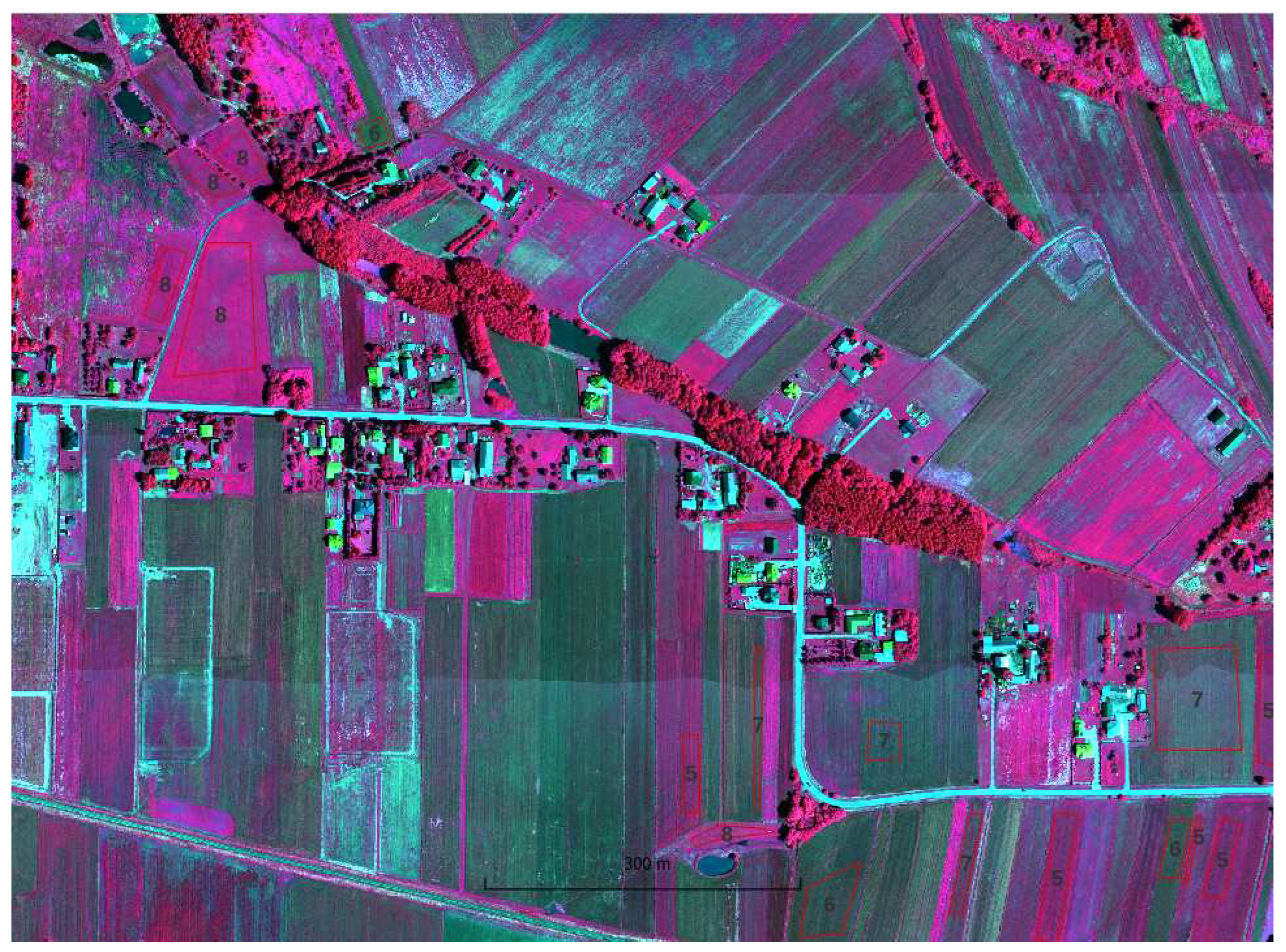
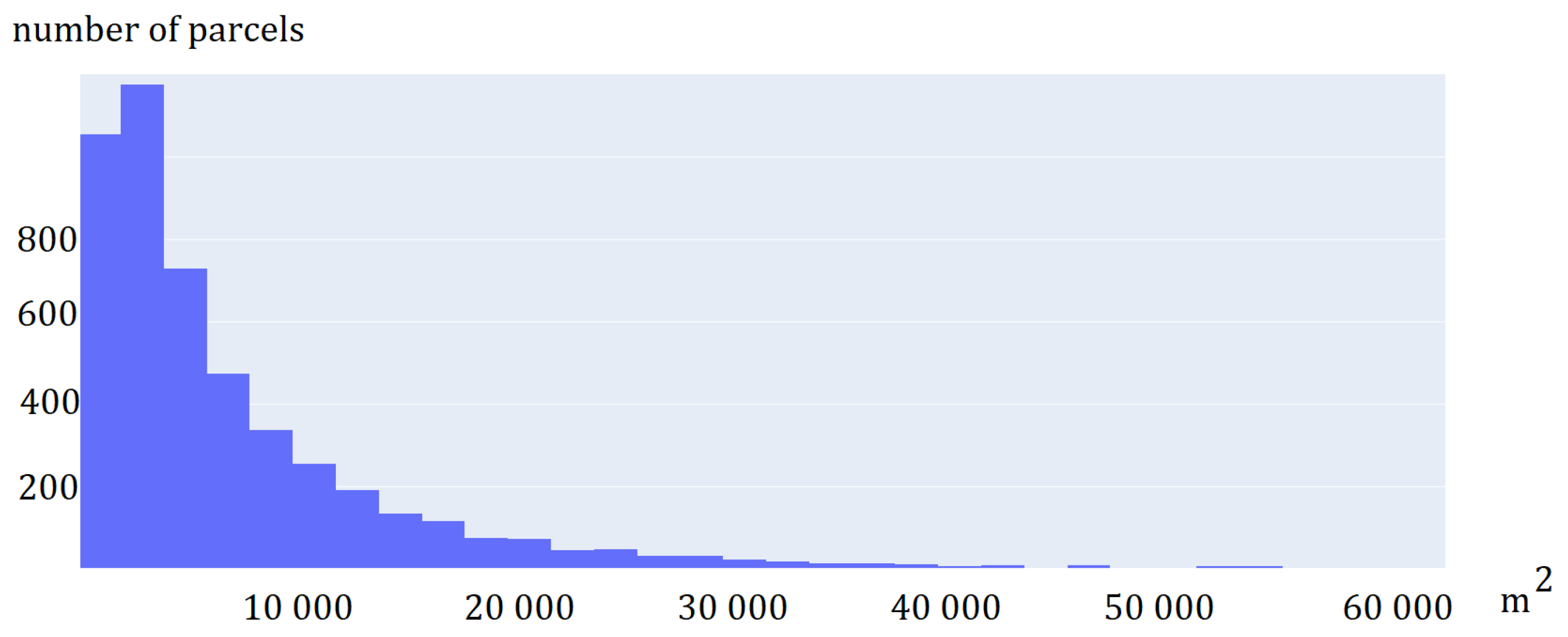
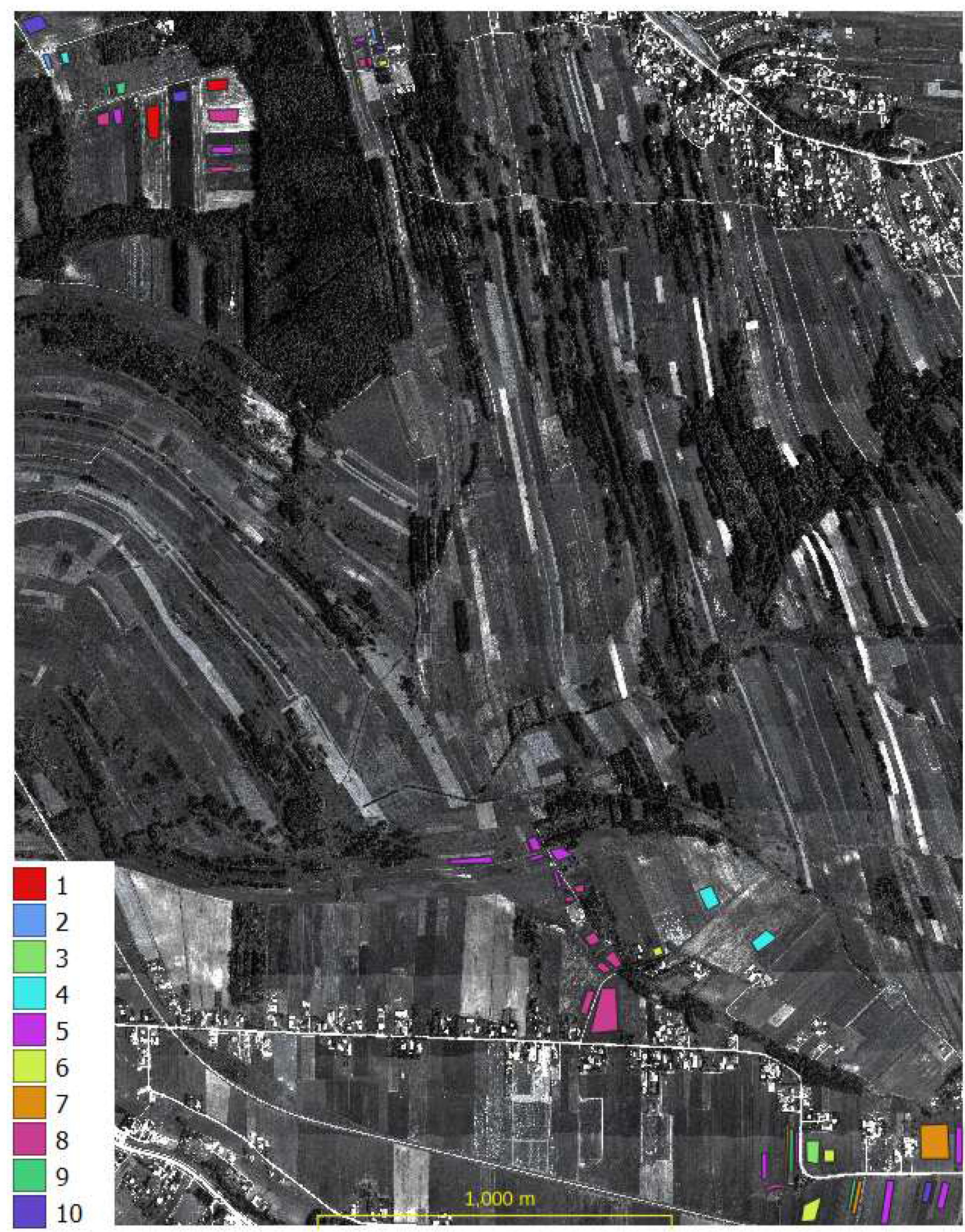
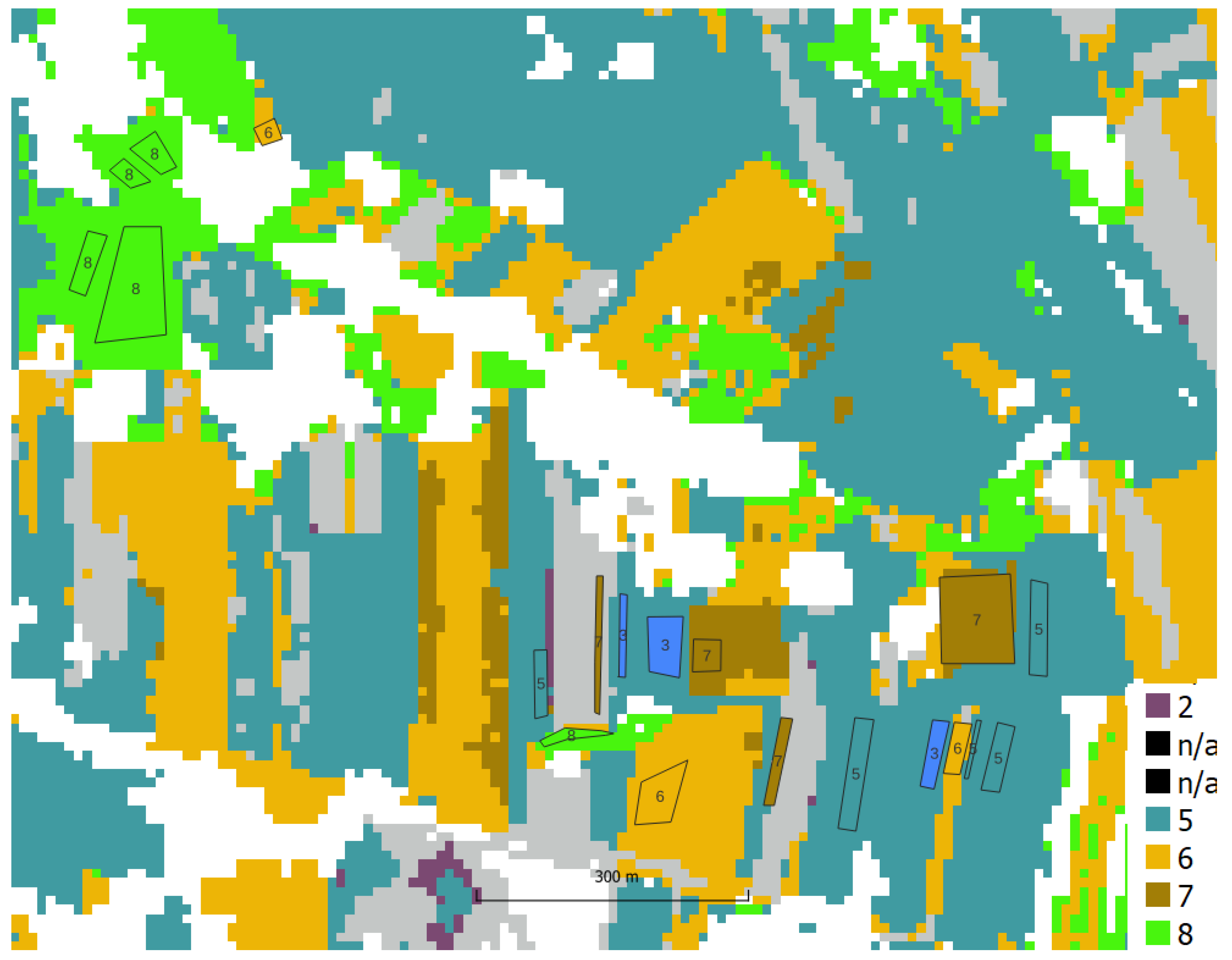
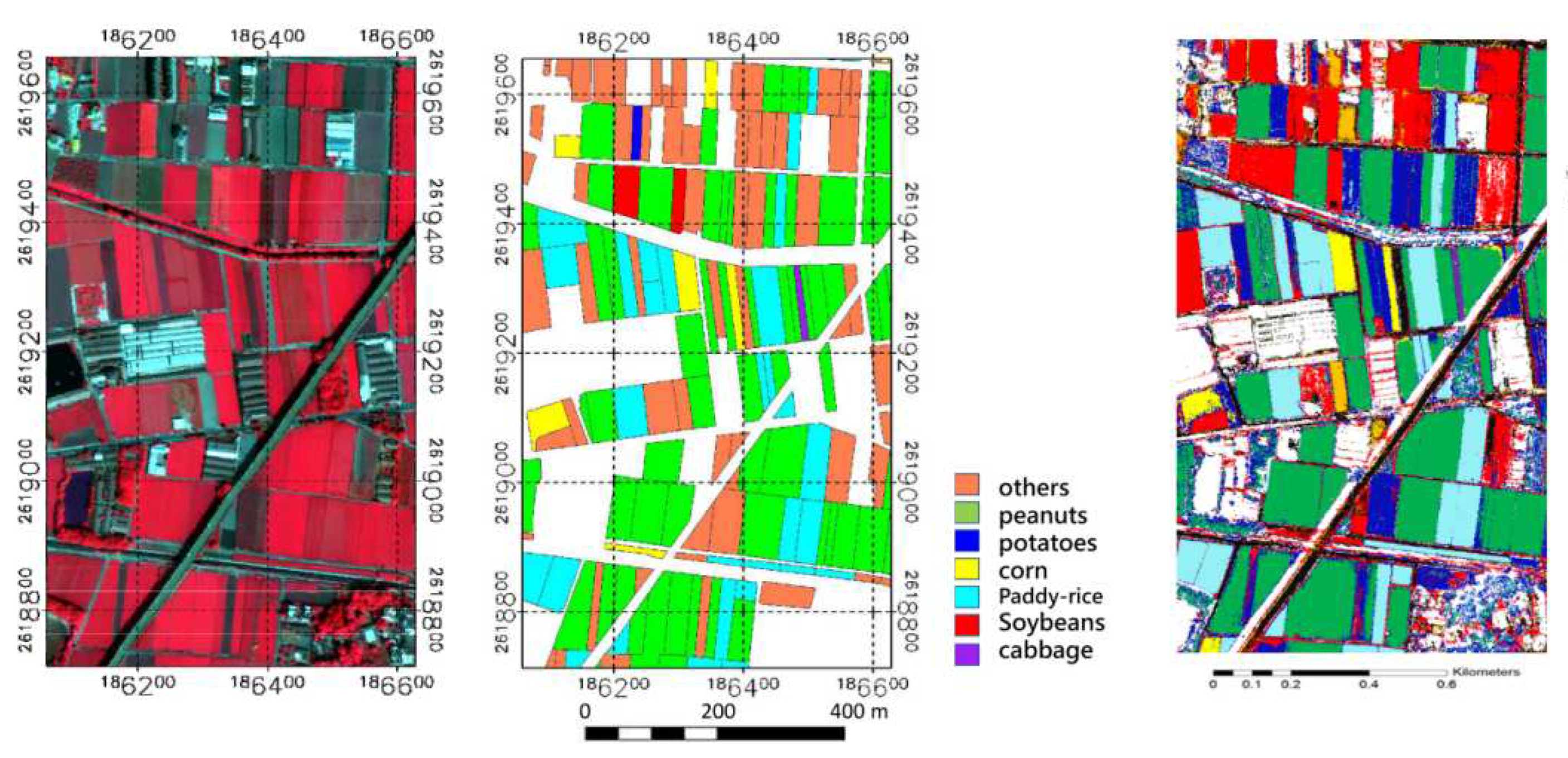
A field visit was conducted to obtain information about the ground truth (the plant that was 2021 grown on the agriculture parcels). Information on 56 agricultural plots was acquired by positioning the location using handheld GPS, 10 classes were defined:
DTM and DSM (Digital Surface Model) were obtained from the national server: geoportal.gov.pl. Three DSM sheets (about 150 megabytes) and 15 DTM sheets (about 160 megabytes) with a pixel size of 1m.
2.3. Data preprocessing
Six Sentinel-2 images were subset and resampled into 10 m. From each Sentinel-2 set 10 bands were selected (B2, B3, B4, B5, B6, B7, B8, B8A, B11 and B12) for area of interest (AOI): 1295 columns x 922 rows (UL: 552250, 5567620 ; LR: 5558400, 5565200 ; EPSG:32634). The channels of all the images prepared in this way were saved in a single TIF file. In addition, NDVI was calculated for each registration date and added to the above mentioned TIF file. Image classification also uses other data that can increase the accuracy of classification, such as numerical terrain models and the slopes/aspects calculated from them. So SRTM was acquired, cropped to the AOI, and resampled to 10m. SRTM and calculated: slope and aspect were added to the TIF file as well. This means that the S2 time series stack consists of 60 Sentinel-2 channels, 6 NDVI images and 3 images containing topography information of the area.
The original Sentinel-2 images are recorded on 12 bits and stored on 16 bits as uint16 (in the metadata there is a size by which DN should be divided to calculate the reflection coefficient: 0-1 as a float32 number, which is 10,000). The values of NDVI coefficients change from -1 to 1 and were stored as float32 numbers.
The SRTM layer is of float32 type and includes for AOI values in the range of 230-310m, slope and aspect are also float 32 type and include values in the range of: 0 to 26 degrees, and 0 to 360 degrees. For the purposes of machine learning, all layers were scaled to a range: 0.0 - 1.0.
Due to the large size of the hyperspectral image, it was cropped of the area where the field visit was conducted and resampled to a pixel size of 1m and 3m. Numerical terrain models were merged and clipped to the extent of the hyperspectral image. In addition, slopes and aspects were calculated from the DTM and NDVI from hiperspectral channels. All rasters were merged into a single TIFF file (5340 cols x 6840 rows, UL: 557062, 5566510 ; LR: 559732, 5563090, 430 bands, NDVI, DTM, DSM, slope and aspect).
The hyperspectral mosaic (9484 rows x 7478 cols, UL: 554995, 5566821 ; LR: 559737, 5563082) made from the processed hyperspectral images has a spatial resolution of 0.5 m, consists of 430 spectral channels (414.13 nm - 2357.43 nm) is registered in the UTM 34N coordinate system (EPSG:32634) and takes up about 60 gigabytes
2.4. Methods
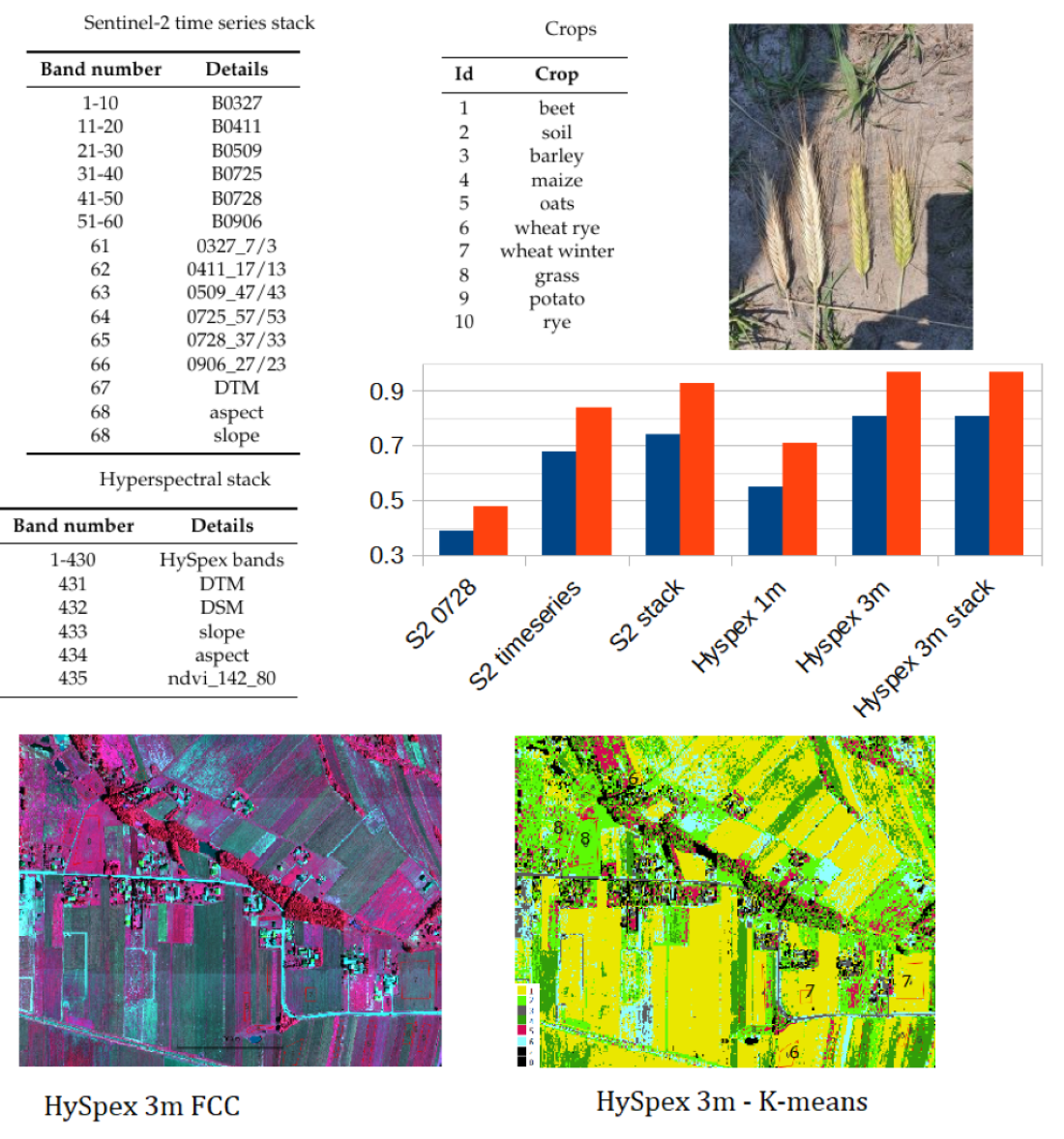
The image data, numerical terrain models and their derivatives were merged using own code in Python as a stack and saved as a single tif file. Separately, one file from the Sentinel-2 time series, at 10m resolution, and one file with hyperspectral data at 3m resolution (the original HySpex 0.5 data was resampled to 3m). There are 68 layers (bands) in the Sentinel-2 time series stack file,
Table 2. From 1-60 Sentinel-2 channels, 61 to 66 NDVI for each date (the channels used for calculation are also given), 67-68 DTM, aspect and slope. There are 435 layers (bands) in the Hyperspectral stack file
Table 3. From 1-430 HySpex channels, 431-434 DTM, DSM aspect and slope, 435 NDVI (the channels used for calculations are also given).
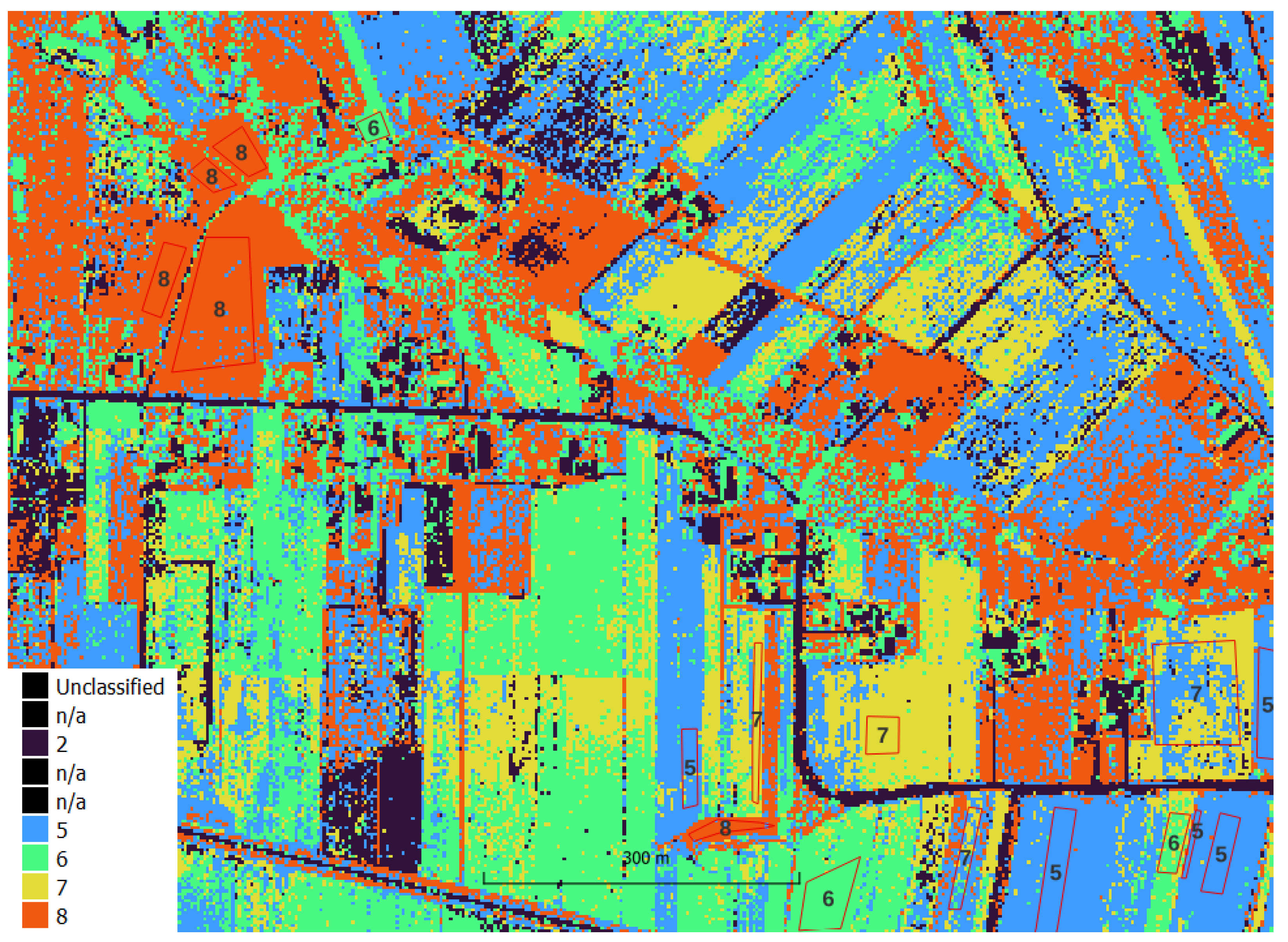
Image processing was carried out using custom scripts in Python and free plug-ins for QGIS (EnMAP-Box and GRASS). Automatic classification was performed in an unsupervised (K-means method) and supervised (Random Forest method) manner. RF classification accuracy analysis was analyzed by k-fold cross validation in EnMAP-Box. Analysis of the accuracy of the final classification result was done on independent test fields in GRASS.
Clustering in EnMAP proceeds in two stages: FitKMeans and Predict Clustering
FitKMeans in EnMAP-Box is executed using the following script:
Argument of class - "‘k-means++’ : selects initial cluster centroids using sampling based on an empirical probability distribution of the points’ contribution to the overall inertia" (scikit-learn 1.1.2). Number of clusters can be modified (default = 8).
Another issue is standardize features by removing the mean and scaling to unit variance. The standard score of a sample x is calculated as: z = (x - u) / s where u is the mean of the training samples or zero if with_mean=False, and s is the standard deviation of the training samples or one if with_std=False (scikit-learn 1.1.2).
The second step performs , which applies a to a raster.
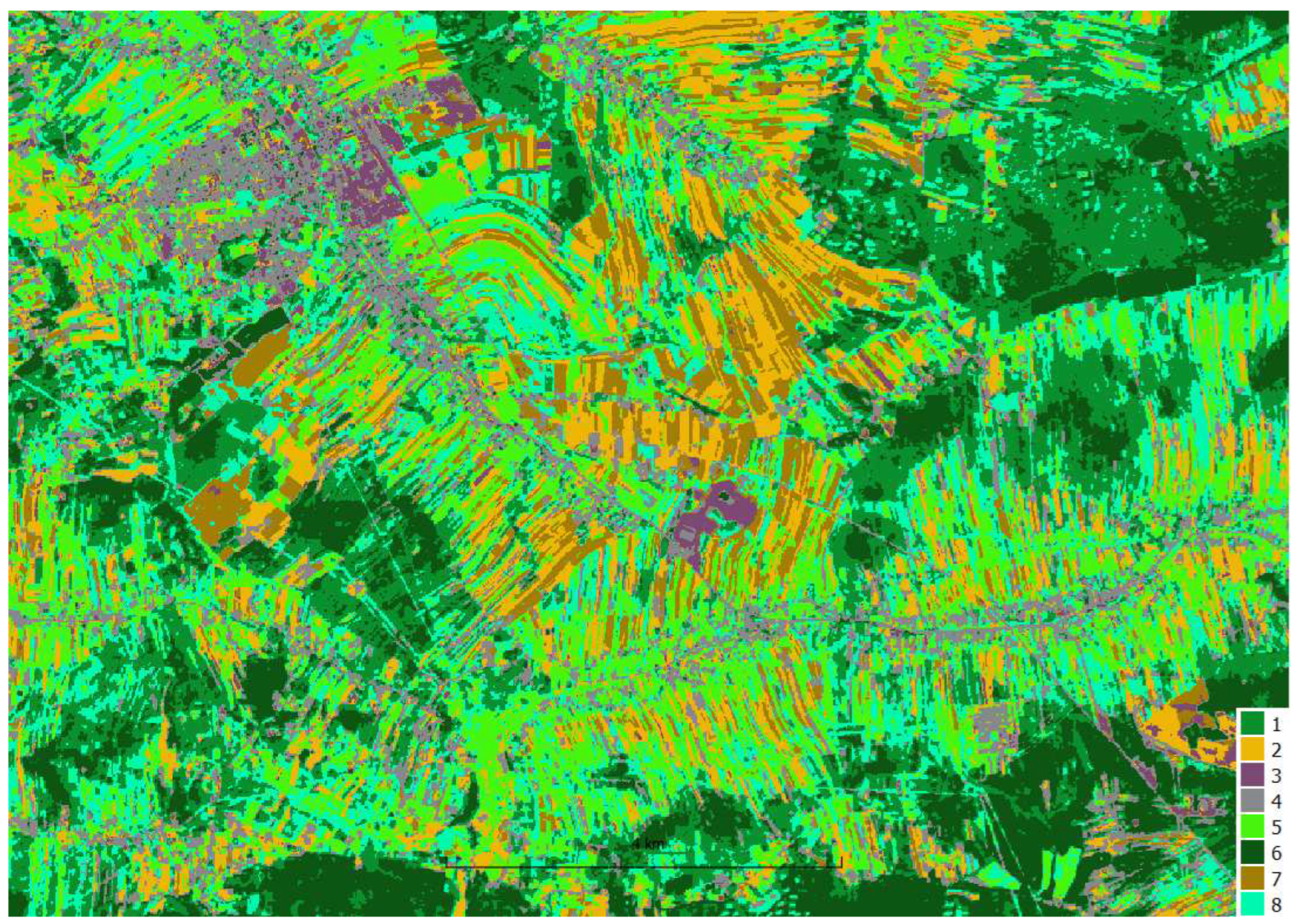
The resulting clusters were mapped to crop type and analyzed in the context of reference fields to evaluate the effectiveness of the method.
Random Forest in EnMAP in classic approach proceeds using the following script:
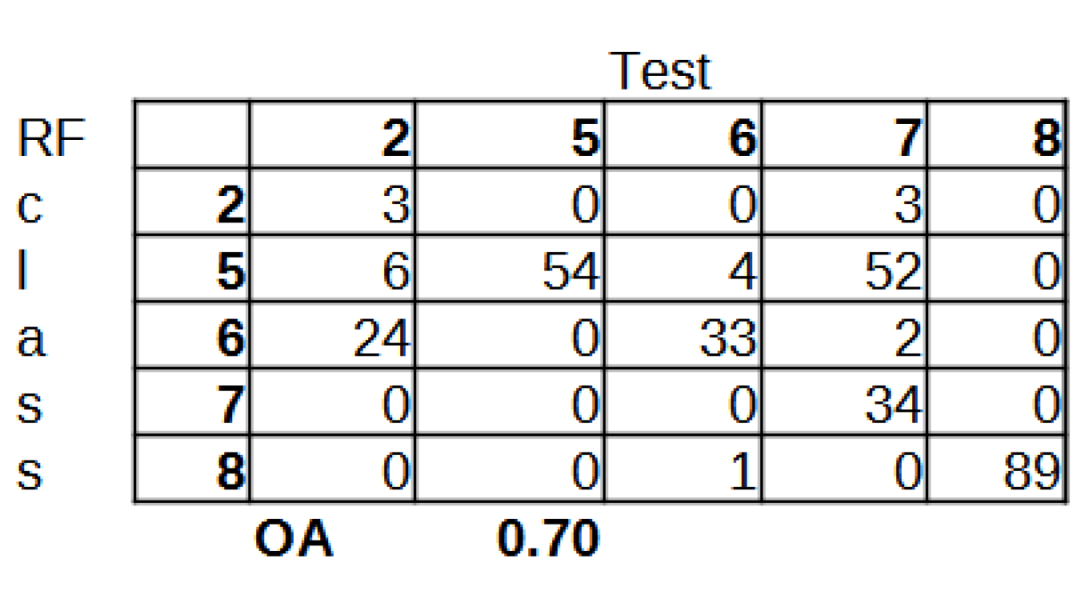
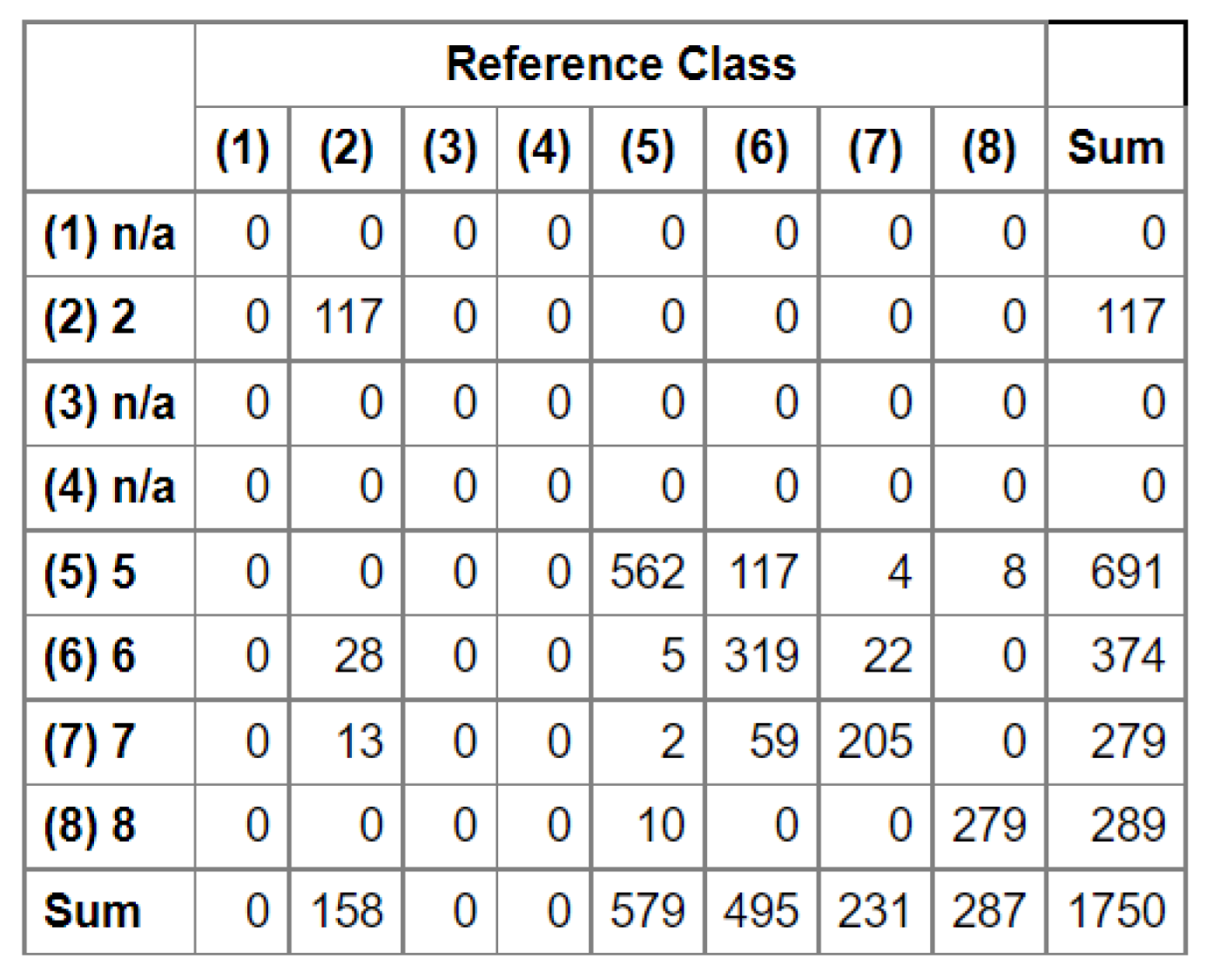
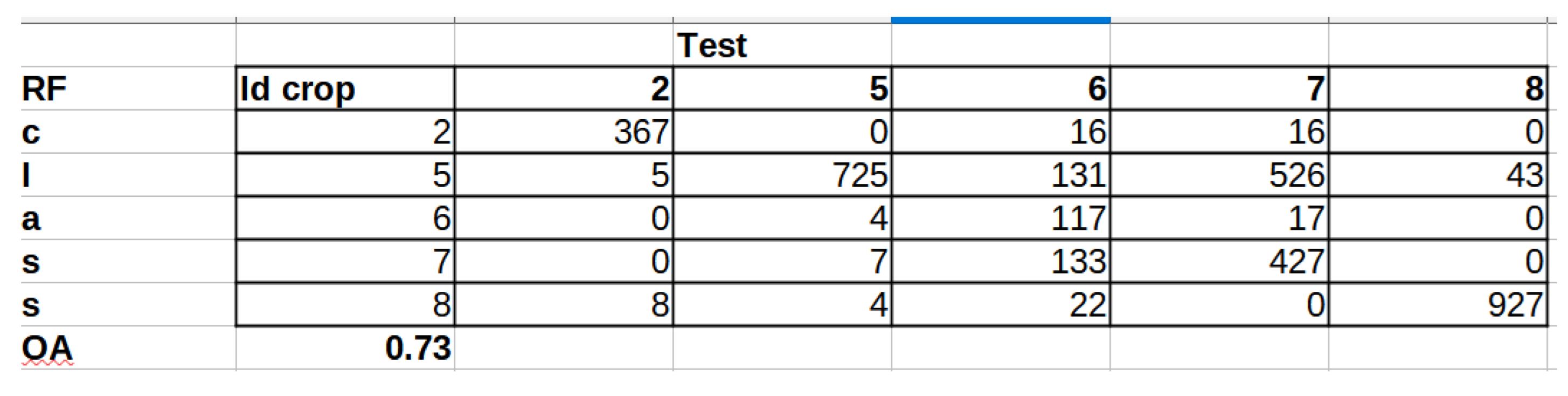
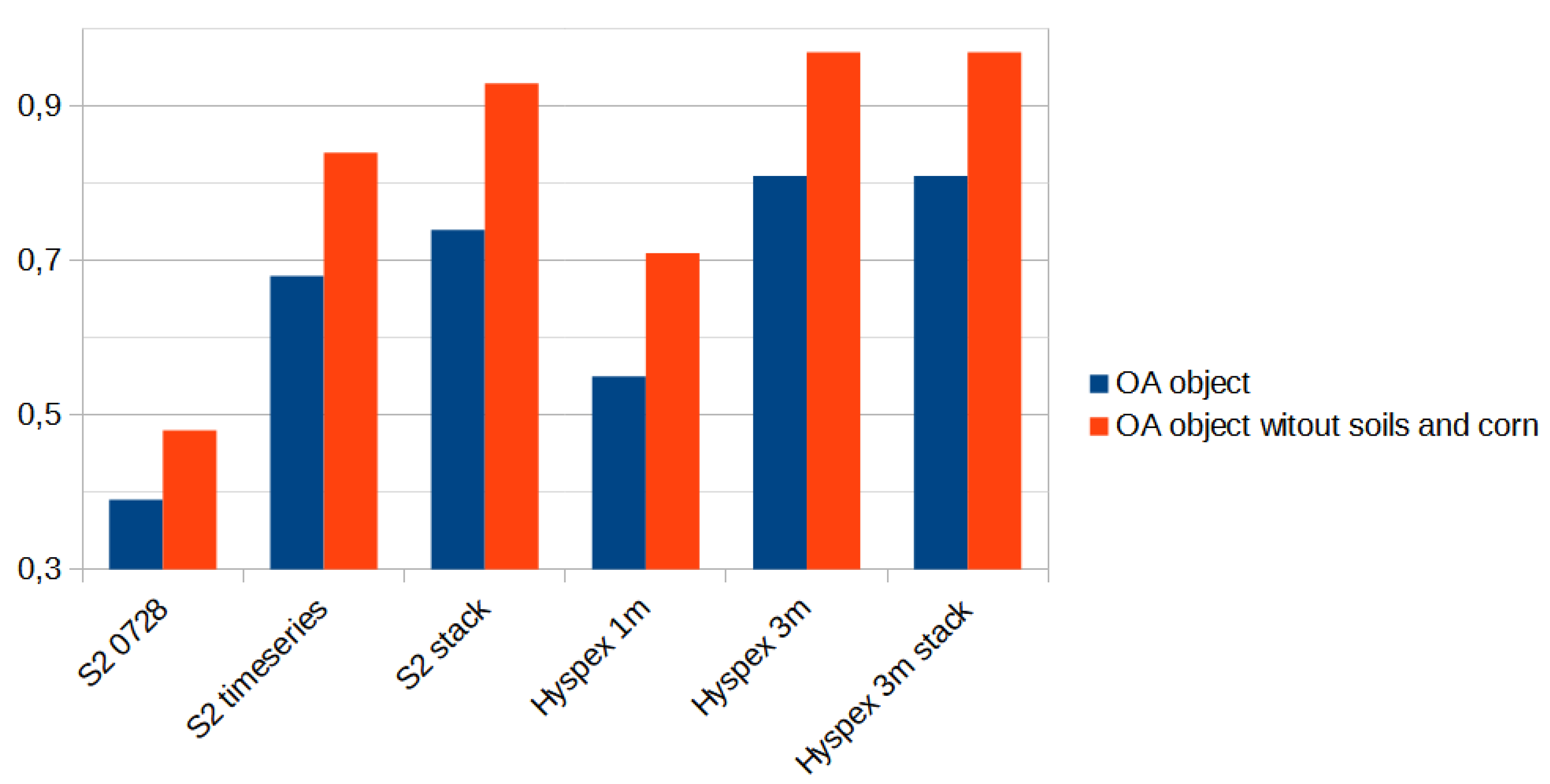
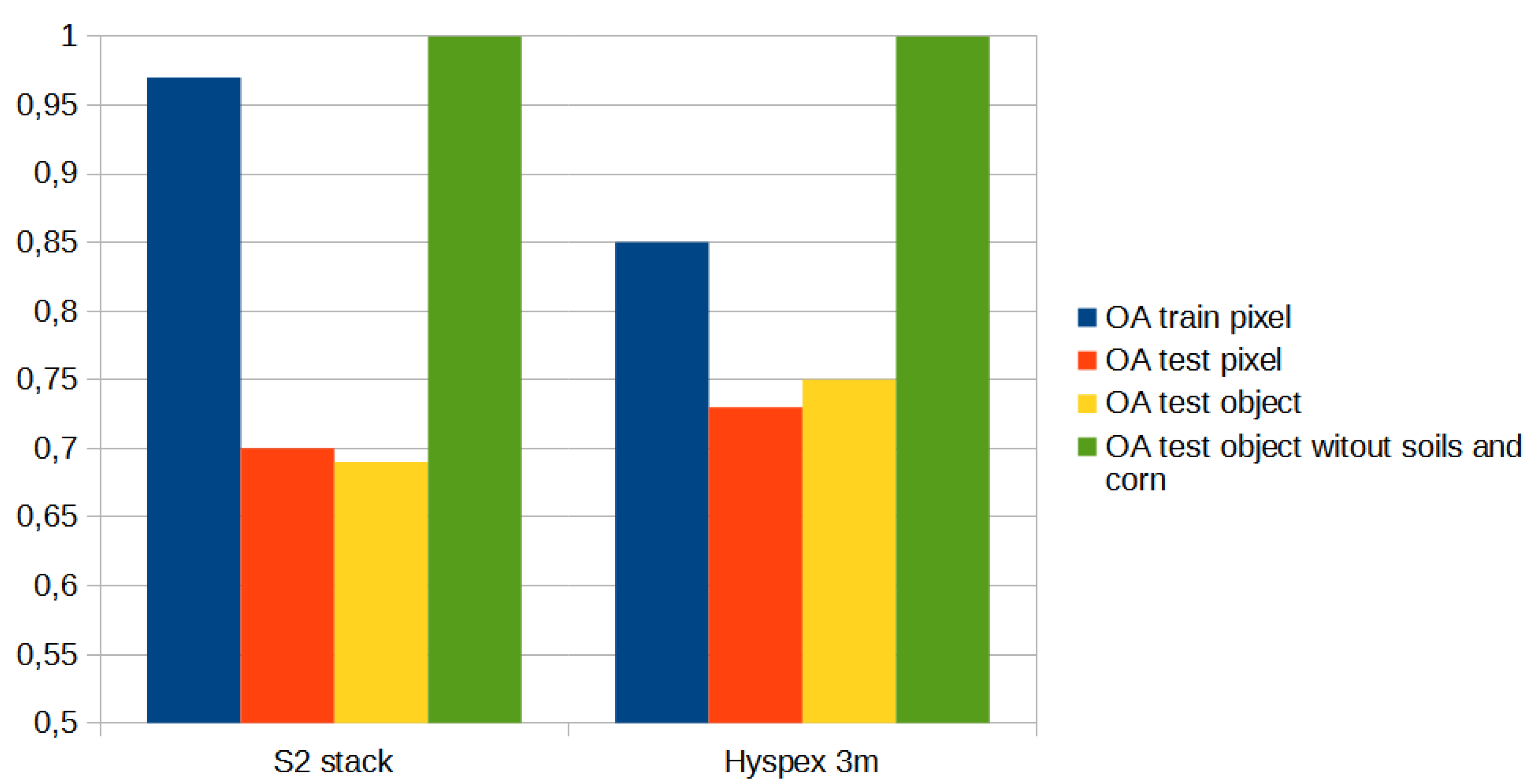
The set of reference parcels was divided into two separate sets using stratified random sampling. The learning process was based on the training set. The model’s accuracy, known as validation accuracy, was determined using k-fold cross-validation (defaulting to 3 folds). To select the optimal hyperparameters for our Random Forest (RF) model, we utilized the widely used grid search method through scikit-learn’s Grid Search CV class. Our training phase employed three evaluation metrics - accuracy, balanced accuracy (mean recall), and f1-weighted (weighted average of precision and recall) - to measure the model’s performance. After testing various configurations using 10-fold cross-validation, we ultimately settled on the following classification settings.
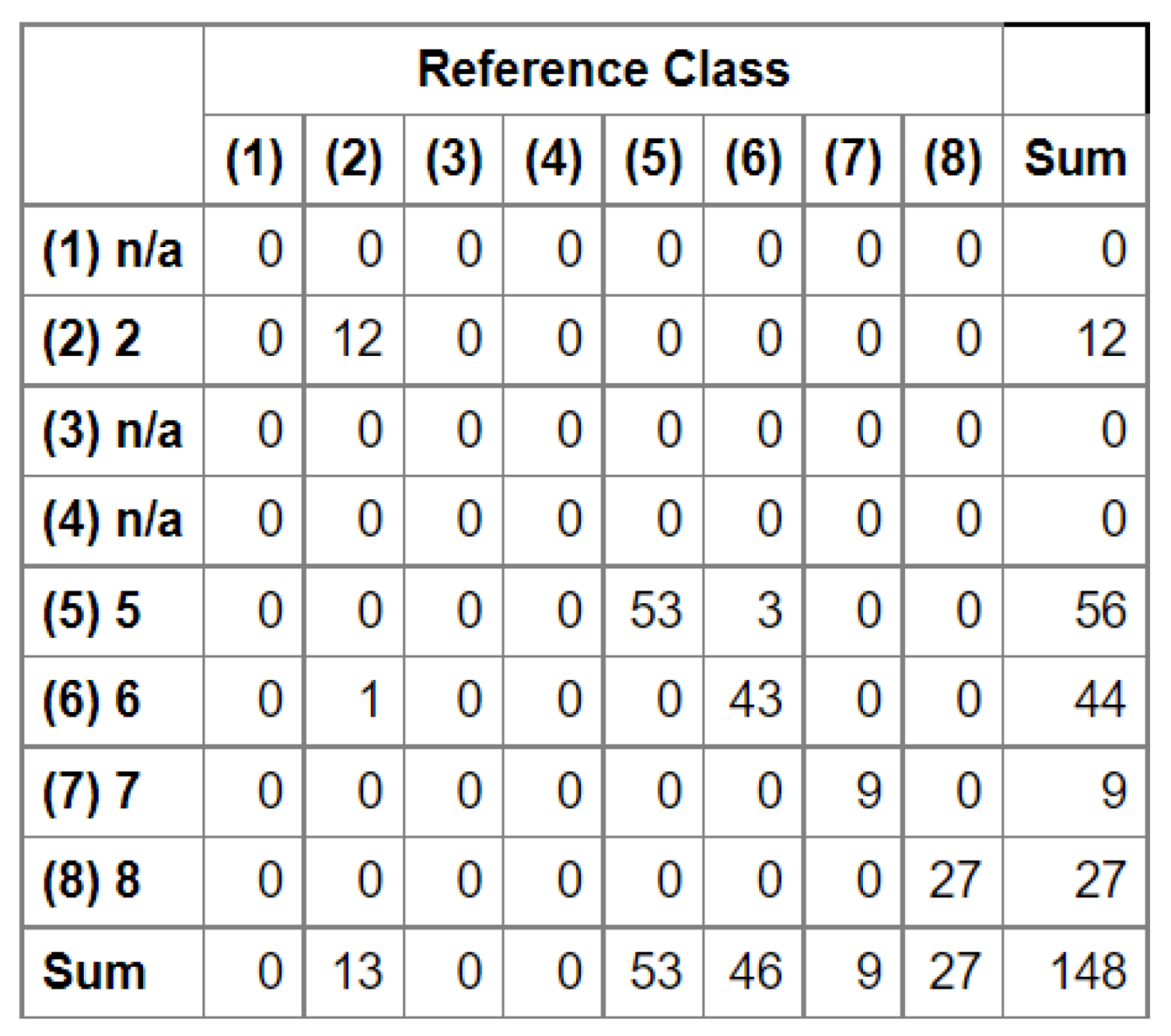
In the next stage, we conducted an accuracy analysis based on a test set that was not involved in the learning process. Although accuracy analysis on independent test fields is available in EnMAP-Box, the
function from the GRASS plugin was used for technical reasons. Function "
tabulates the error matrix of classification result by crossing classified map layer with respect to reference map layer. Both overall kappa (accompanied by its variance) and conditional kappa values are calculated. This analysis program respects the current geographic region and mask settings" (
https://ibiblio.org/pub/packages/gis/grass/grass63/manuals/html63_user/r.kappa.html). In this manner, pixel accuracy was determined. In addition, accuracy was analyzed at the plot level by performing an automatic majority class extraction for each polygon (QGIS-Processing tools-Zonal statistic).
Even though researchers have investigated different metrics over the years [
26,
27,
28,
29,
30,
31]), the most commonly recommended metric remains Overall Accuracy (OA) [
32], which is defined as OA = TP / (TN + FP + FN) [
28]. Therefore, in our study, we limited our analysis to OA.
The study aimed to compare the accuracy of classification results from Sentinel-2 and hyperspectral images captured at significantly different altitudes (10 m for satellites and 1-3 m for aerial images). Processing of single registrations from both types of images as well as time series of Sentinel-2 was conducted to achieve this goal. The complete dataset (stack) was used for classification, and the accuracy was compared to the classification results obtained by excluding NDVI, DEM, and their processing from the remote sensing data.