Submitted:
01 February 2023
Posted:
03 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. How to use this guide
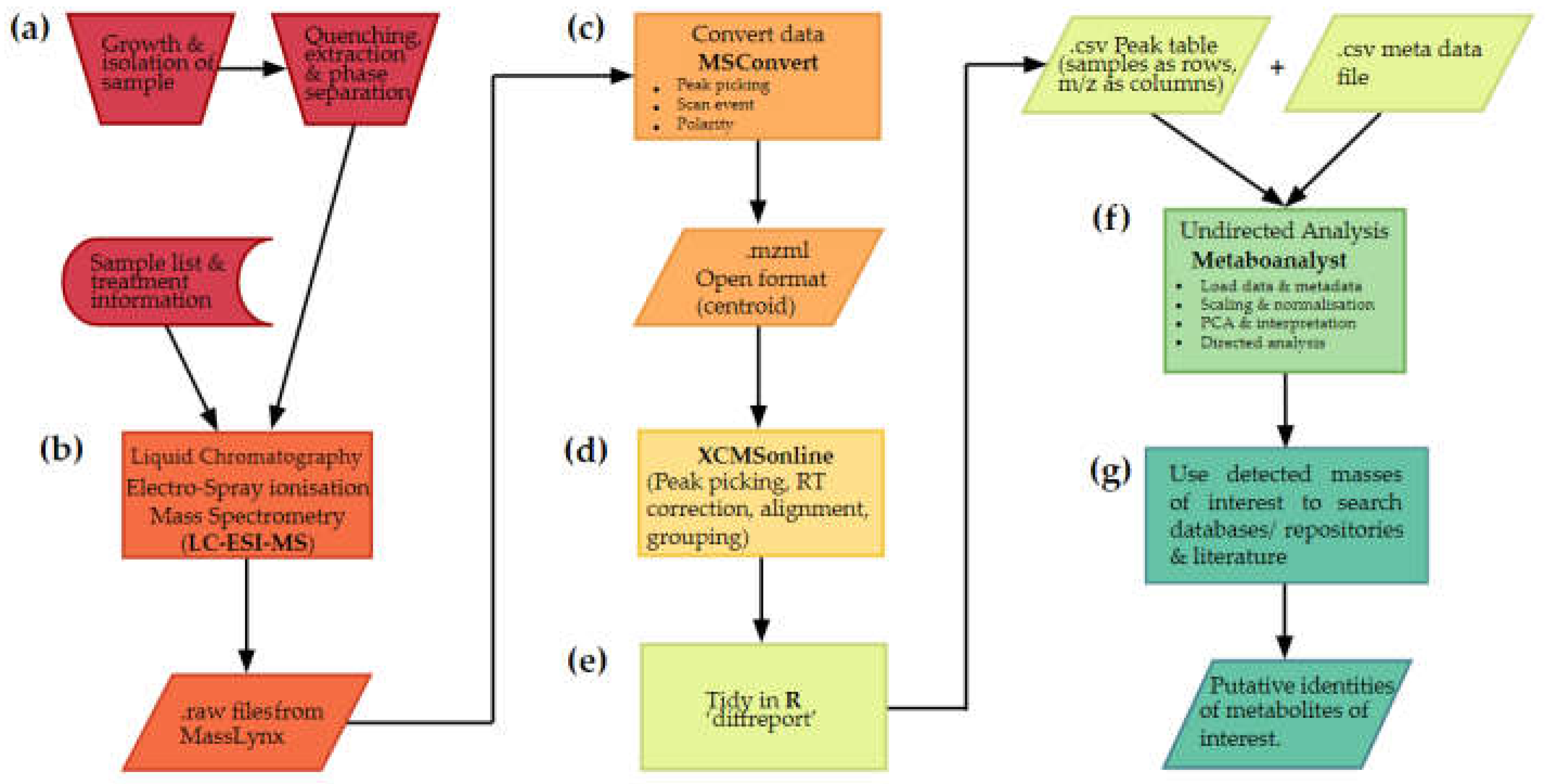
2.1.1. Stages of an untargeted metabolomics workflow
2.1.3. Assumptions
- a basic understanding of R and RStudio, and of metabolomics technologies in general (experienced R users are also encouraged to consider tools such as the RforMassSpectrometry initiative https://rformassspectrometry.org accessed on 27 January 2023);
- access to the internet and remote access to raw data;
- used Waters mass spectrometers and MassLynx software to obtain data (or equivalent steps and outcomes from other instruments);
- access to a sample list from MassLynx and treatment information (i.e. metadata, though our code can help format this);
- files with unique identifiers, ideally in the format of the following example:
- experiment-identifier_001.raw
- experiment-identifier_002.raw
- experiment-identifier_003.raw
2.1.4. Experimental structure
- case vs control
- wild-type vs transgenic line
- Strain 1 vs. strain 2 vs. strain 3
- Two factors with two or more levels in each such as +/- treatment for two strains
- Time course for one or two factors such as +/- treatment for two strains over three time points
- What are the biological replicates being analysed and are they independent of each other (or has the same organism/ population been sampled multiple times)?
- Are there technical replicates (i.e. repeated runs of the same sample)?
- Are Quality Control (QC) samples required? Are analytical standards needed? (See box 2.1.5)
- What groupings are required to answer the research questions outlined?
2.1.5. Quality Control
- Spike all prepared samples with a compound for which the m/z (and RT) is known and which is unlikely to be otherwise present in the experimental samples;
- Prepare a pooled QC sample from an aliquot of each of the samples and include this at regular intervals in the MS run;
- Include blanks and/ or extraction blanks at regular intervals in the MS run;
- Use lock mass calibration (for Waters instruments).
- Check file sizes of .raw files across the MS run;
- Check file sizes of converted .mzML files - reconvert any that are unexpected;
- Compare spectra between technical replicates
2.1.6. Nice, neat metadata for analysis
- “Filename”: this is a list of the filenames of the .mzml files (the part before the .mzml)
- “Filetext”: this is the name that has been manually added to the metadata of that sample in MassLynx (this can be found at $$ SampleID: in the _HEADER.txt file of the original .RAW folder if it is not already known)
- “MSFile” or an equivalent column that contains either “pos” or “neg” within it Any other columns will be ignored in this file
- “Filetext”: this must contain all the distinct values of “Filetext” from
- samplelist.csv
- “Variable1”: the naming of this column is left to the user (but spaces are to be avoided: instead use “-” or “_"). For example, in an MS run comparing a wild-type to a control for example, this column could be named “treatment” and filled with “WT” and “C” as appropriate
- “Variable2” etc: further variables. This may include batch identifiers (for example if many samples were run over multiple days), treatments or environmental variables
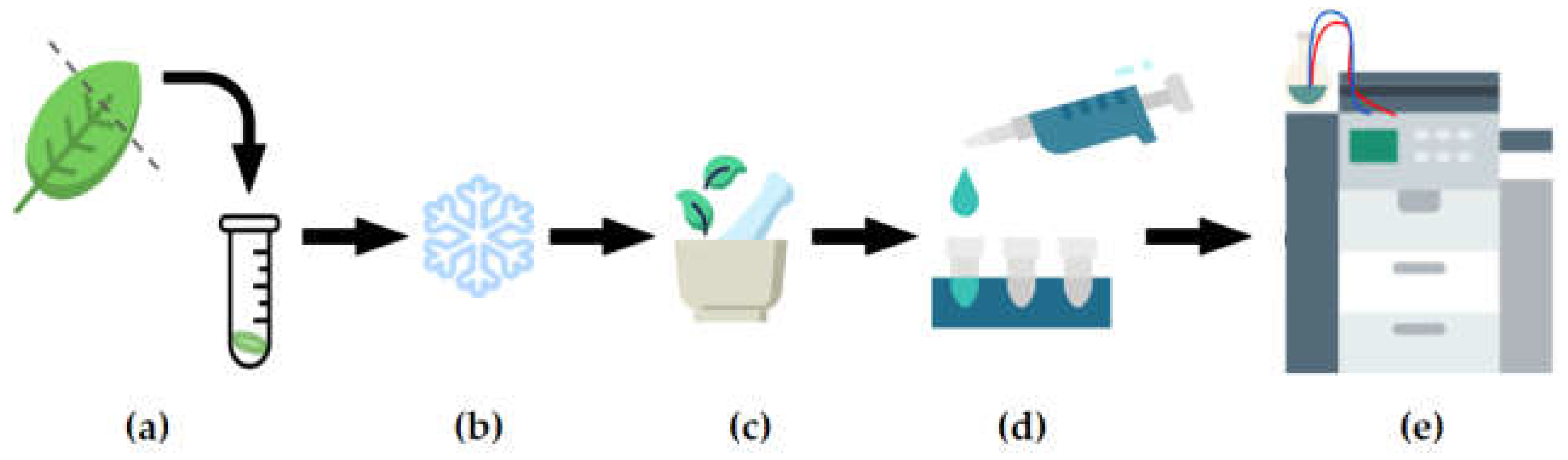
2.2. Metabolite extraction and data acquistion
2.3. Converting data to open format using Proteowizard
2.4. Preprocessing data
- baseline correction and/ or noise reduction (estimating what part of the detected intensity is the sample and “cleaning away” or adjusting the spectra to show only the signal believed to be associated with the sample);
- normalisation and/ or standardisation (these can mean a range of different things to different people but broadly cover accounting for differences in sample volume or concentration or total intensity of the signal);
- grouping and peak picking (wave-form algorithms are used to determine which parts of the spectra constitute separate peaks utilising their m/z value);
- alignment or peak matching (assessing across samples to determine whether peaks with slightly different m/z values are the same peak so that samples can be compared more reliably).
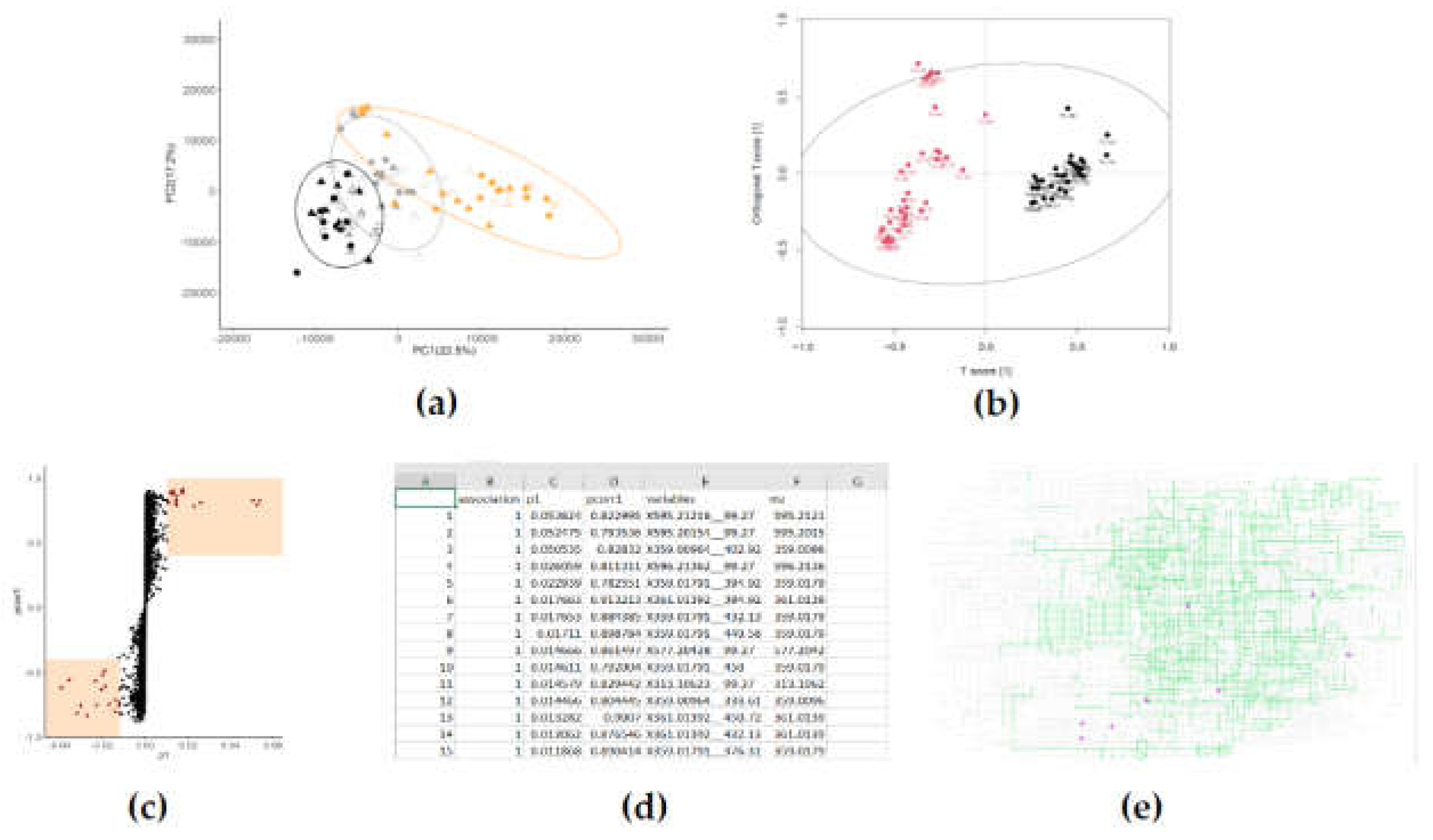
2.5. Multivariate analysis
- Are the metabolomic fingerprints distinct classes (treatment groups) different from each other?
- Which features of the metabolomic fingerprint are causing them to be different from each other?
2.6. What are my metabolites?
- METLIN to search by m/z;
- KEGG PATHWAY and KEGG COMPOUND [31] to corroborate likelihood of detecting certain compounds in the study organism/ sample and to gain insight on biological function;
- Data repositories such as MetaboLights;
- Reporting Metabolomics Standards Initiative (MSI) identification levels (see also [36]).
2.7. Sharing metabolomics data
- Findable
- Accessible
- Interoperable
- Reusable
2.7.1. MetaboLights repository
2.8. Citation of the tools used in the workflow
- Cite all R packages used in our functions (see function at start of each R code to produce list of references)
- Use citation(R) and RStudio.Version() to get the version information for R and RStudio that you have used for analysis;
- Proteowizard (SeeMS and MSConvert) citation;
- Up to date Metaboanalyst citation;
- Up to date XCMS online and METLIN citations;
- MassUp citation;
- MassBank citation (include access date);
- Up to date ECMDB citation (don’t forget any other organism specific metabolite databases used);
- Up to date KEGG citation (including BRITE, COMPOUND and PATHWAY);
- Up to date PubChem citation;
- Write a data availability statement in any publication that links to your archived data in MetaboLights.
Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allwood, J.W.; Williams, A.; Uthe, H.; van Dam, N.M.; Mur, L.A.J.; Grant, M.R.; Pétriacq, P. Unravelling Plant Responses to Stress—The Importance of Targeted and Untargeted Metabolomics. Metabolites 2021, 11(8), 558. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Cravatt, B.F.; Siuzdak, G. The expanding role of mass spectrometry in metabolite profiling and characterization. ChemBioChem 2005, 6(11), 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Vincent, I.M.; Ehmann, D.E.; Mills, S.D.; Perros, M.; Barrett, M.P. Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrobial Agents and Chemotherapy 2016, 60(4), 2281–2291. [Google Scholar] [CrossRef] [PubMed]
- Di Minno, A.; Gelzo, M.; Stornaiuolo, M.; Ruoppolo, M.; Castaldo, G. The evolving landscape of untargeted metabolomics. Nutrition, Metabolism and Cardiovascular Diseases 2021, 31(6), 1645–1652. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jasbi, P.; Shi, X.; Turner, C.; Hrovat, J.; Liu, L.; Rabena, Y.; Porter, P.; Gu, H. Early Breast Cancer Detection Using Untargeted and Targeted Metabolomics. J. Proteome Res 2021, 20, 3133. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. Journal of the American Society for Mass Spectrometry 2016, 27(12), 1897–1905. [Google Scholar] [CrossRef]
- Dudzik, D.; Barbas-Bernados, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. Journal of Pharmaceutical and Biomedical Analysis 2018, 147, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Vicini, A.; Salzer, L.; Stanstrup, J.; Badia, J.M.; Neumann, S.; Stravs, M.A.; Verri Hernandes, V.; Gatto, L.; Gibb, S.; Witting, M. A Modular and Expandable Ecosystem for Metabolomics Data Annotation in R. Metabolites 2022, 12, 173. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8(2), 31. [Google Scholar] [CrossRef]
- Misra, B.B. New tools and resources in metabolomics: 2016–2017. Electrophoresis 2018, 39(7), 909–923. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Current Opinion in Biotechnology 2019, 55, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Jorge, T.F.; Mata, A.T.; António, C. Mass spectrometry as a quantitative tool in plant metabolomics. Philos Trans A Math Phys Eng Sci 2008, 374(2079), 20150370. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Su, X; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annual Review of Biochemistry 2017, 86(1), 277–304. [Google Scholar] [CrossRef] [PubMed]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; González-Ruiz, V.; Rudaz, S. Implementation of liquid chromatography-high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: a tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef]
- Villate, A.; San Nicolas, M.; Gallastegi, M.; Aulas, P.-A.; Olivares, M.; Usobiaga, A.; Etxebarria, N.; Aizpurua-Olaizola, O. Metabolomics as a Prediction Tool for Plants Performance under Environmental Stress. Plant Sci. 2021, 303, 110789. [Google Scholar] [CrossRef] [PubMed]
- Austen, N.; Walker, H.J.; Lake, J.A.; Phoenix, G.K.; Cameron, D.D. The Regulation of Plant Secondary Metabolism in Response to Abiotic Stress: Interactions Between Heat Shock and Elevated CO2. Frontiers in Plant Science 2019, 10, 1463. [Google Scholar] [CrossRef]
- Martens, L.; Chambers, M.; Sturm, M.; Kessner, D.; Levander, F.; Shofstahl, J.; Tang, W.H.; Römpp, A.; Neumann, S.; Pizarro, A.D., Montecchi-Palazzi, L.; Tasman, N.; Coleman, M.; Reisinger, F.; Souda, P.; Hermjakob, H.; Binz, P-A.; Deutsch, E.W. mzML—a Community Standard for Mass Spectrometry Data. Mol. Cell. Proteomics 2011, 10(1), R110.000133. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 2008, 24(21), 2534–2536. [Google Scholar] [CrossRef]
- Forsberg, E.; Huan, T.; Rinehart, D.; Benton, H.P.; Warth, B.; Hilmers, B.; Siuzdak, G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat Protoc 2018, 13, 633–651. [Google Scholar] [CrossRef]
- López-Fernández, H.; Santos, H.M.; Capelo, J.L.; Fdez-Riverola, F.; Glez-Peña, D.; Reboiro-Jato, M. Mass-Up: an all-in-one open software application for MALDI-TOF mass spectrometry knowledge discovery. BMC Bioinformatics 2015, 16, 318. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O'Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching and identification. Analytical Chemistry 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Gibb, S.; Strimmer, K. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012, 28(17), 2270–2271. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucl. Acids Res. 2009, 37, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Metaboanalyst tutorials: available at https://dev.metaboanalyst.ca/docs/Tutorials.xhtml (accessed on 27/01/2023).
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; van der Gheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswamy, P.; Teo, G.; Ow, J.R.; Lau, A.; Kaldis, P.; Tate, S.; Choi, H. MetaboKit: a comprehensive data extraction tool for untargeted metabolomics. Mol. Omics 2020, 16, 436. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M., Casagrande, J., Davuluri, R., Eds.; Springer, Boston, MA. 2010; pp. 267-277. [CrossRef]
- Kuhl, C; Tautenhahn, R; Boettcher, C; Larson, T.R.; Neumann, S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Analytical Chemistry 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Research 2020, 48(D1), D440–D444. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Analytical Chemistry 2018, 90(5), 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG Bioinformatics Resource for Plant Genomics and Metabolomics. In Plant Bioinformatics; Methods in Molecular Biology; Edwards, D., Eds. Humana Press, New York, NY, 2016; volume 1374. [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; Oda, Y.; Kakazu, Y.; Kusano, M.; Tohge, T.; et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; Zaslavsky, L.; Zhang, J.; Bolton, E.E. PubChem 2023 update. Nucleic Acids Res. 2023, 51(D1), D1373–D1380. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; Krummenacker, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Research 2014, 42(1), D459–D471. [Google Scholar] [CrossRef] [PubMed]
- The Metabolomics Workbench: available at https://www.metabolomicsworkbench.org/ (accessed on 27/01/2023).
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K.; Roessner, U.; Trengove, R. Proposed quantitative and alphanumeric metabolite identification metrics. Metabolomics 2014, 10, 1047–1049. [Google Scholar] [CrossRef]
- Wilkinson, M.; Dumontier, M.; Aalbersberg, I.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3 2016, 160018. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; Heinemann, M.; Link, H.; Luo, J.; Neumann, S.; Nielsen, J.; Perez de Souza, L.; Saito, K.; Sauer, U.; Schroeder, F.C.; Schuster, S.; Siuzdak, G.; Skirycz, A.; Sumner, L.W.; Snyder, M.P.; Tang, H.; Tohge, T.; Wang, Y.; Wen, W.; Wu, S.; Xu, G.; Zamboni, N.; Fernie, A.R. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




