Submitted:
22 January 2023
Posted:
26 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Regulatory Transcription Factors in Cryptococcal Cells
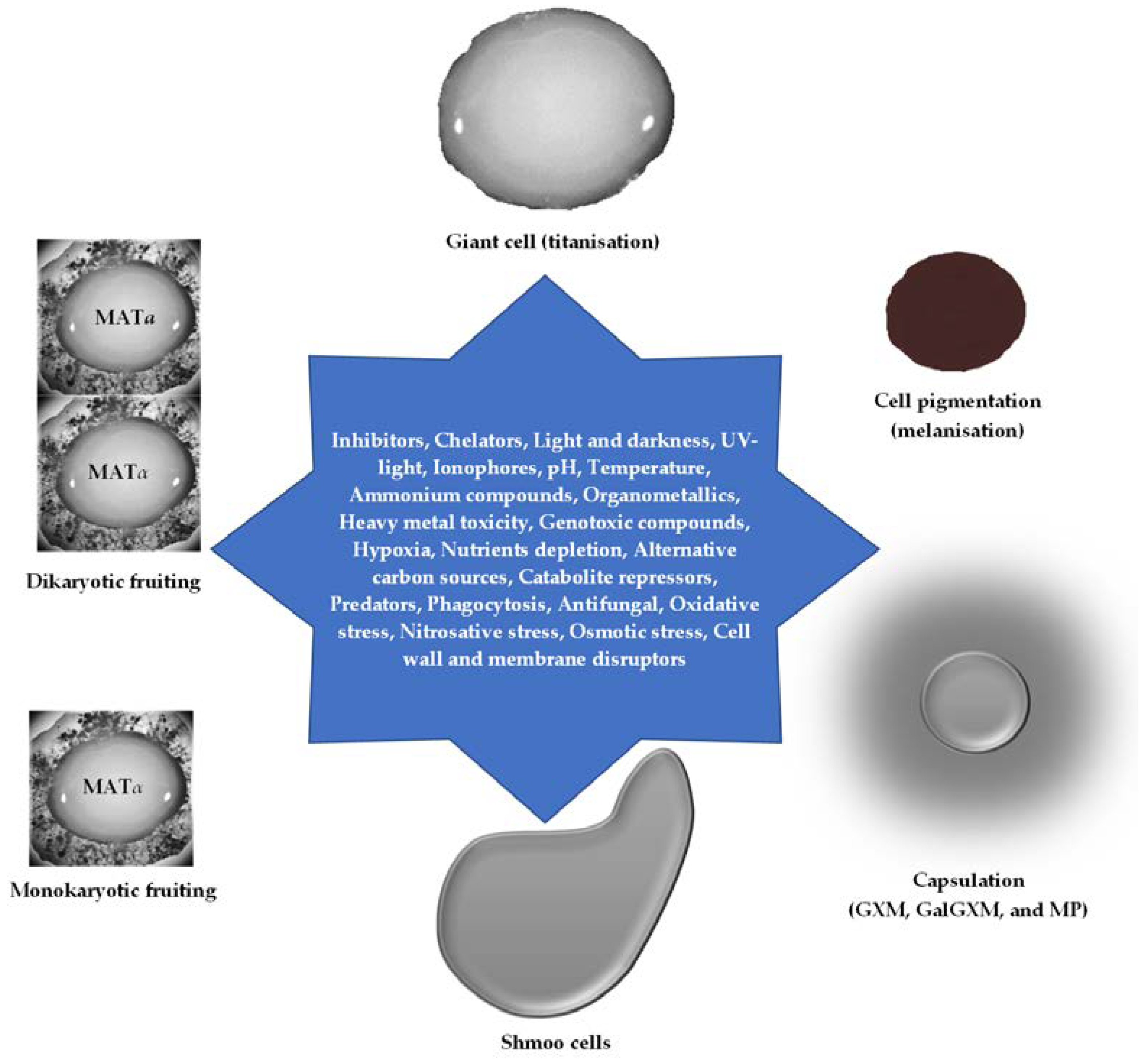
3. Regulatory Transcription Factors in Cryptococcal Cells Control Phenotypic Expression for Morphotypes, Adaptation, Survival, and Virulence
3.1. Cell Wall Chitin-Chitosan Components
3.2. Cell Morphotypes and Aneuploidy
3.3. Capsule: Capsular Polysaccharides and Glycoproteins
3.4. Melanin
3.5. Heterokaryotic Mating/Conjugation, Filamentation, and Sporulation/Haploid Fruiting
3.6. Cell Wall and Membrane Integrity: Sensitivity to Temperature, Radiation/Light, Salinity, pH, Antifungal, Genotoxicants, Reactive Radicals (Oxidative and Nitrosative Stress), and Quorum-Sensing Molecules
3.7. Osmotic Shock
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feldmesser, M.; Kress, Y.; Novikoff, P.; Casadevall, A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 2000, 68, 4225–4237. [Google Scholar] [CrossRef] [PubMed]
- Elhariri, M.; Hamza, D.; Elhelw, R.; Refai, M. Eucalyptus Tree: A Potential Source of Cryptococcus neoformans in Egyptian Environment. Int J Microbiol 2016, 2016, 4080725. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, M.; D'Amicis, R.; Zani, A.; Montagna, M.T.; Caggiano, G.; De Giglio, O.; Balbino, S.; De Donno, A.; Serio, F.; Susever, S. Environmental distribution of Cryptococcus neoformans and C. gattii around the Mediterranean basin. FEMS yeast research 2016, 16. [Google Scholar] [CrossRef]
- Ergin, C.; Sengul, M.; Aksoy, L.; Dogen, A.; Sun, S.; Averette, A.F.; Cuomo, C.A.; Seyedmousavi, S.; Heitman, J.; Ilkit, M. Cryptococcus neoformans Recovered From Olive Trees (Olea europaea) in Turkey Reveal Allopatry With African and South American Lineages. Front Cell Infect Microbiol 2019, 9, 384. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Casadevall, A. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect 2003, 5, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Evolution of intracellular pathogens. Annu Rev Microbiol 2008, 62, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Shuman, H.A.; Casadevall, A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 2001, 98, 15245–15250. [Google Scholar] [CrossRef]
- Derengowski Lda, S.; Paes, H.C.; Albuquerque, P.; Tavares, A.H.; Fernandes, L.; Silva-Pereira, I.; Casadevall, A. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell 2013, 12, 761–774. [Google Scholar] [CrossRef]
- Casadevall, A. Amoeba provide insight into the origin of virulence in pathogenic fungi. In Recent advances on model hosts; Springer: 2012; pp. 1-10. [CrossRef]
- Steenbergen, J.N.; Nosanchuk, J.D.; Malliaris, S.D.; Casadevall, A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun 2003, 71, 4862–4872. [Google Scholar] [CrossRef]
- Paul, C.; Emeka, N. Pathogenicity of Cryptococcus neoformans VNI (ST 32) recovered from environmental and clinical isolates in Nigeria. Comparative Clinical Pathology 2019, 28, 1013–1024. [Google Scholar] [CrossRef]
- Litvintseva, A.P.; Kestenbaum, L.; Vilgalys, R.; Mitchell, T.G. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 2005, 43, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Thakur, R.; Vilgalys, R.; Mitchell, T.G. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 2006, 172, 2223–2238. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Mitchell, T.G. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infection and immunity 2009, 77, 3188–3195. [Google Scholar]
- Casadevall, A.; Rosas, A.L.; Nosanchuk, J.D. Melanin and virulence in Cryptococcus neoformans. Curr Opin Microbiol 2000, 3, 354–358. [Google Scholar] [CrossRef]
- Upadhya, R.; Baker, L.G.; Lam, W.C.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis. mBio 2018, 9, e02087–02018. [Google Scholar] [CrossRef]
- Fan, W.; Kraus, P.R.; Boily, M.J.; Heitman, J. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 2005, 4, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Goulart, L.; Rosa e Silva, L.K.; Chiapello, L.; Silveira, C.; Crestani, J.; Masih, D.; Vainstein, M.H. Cryptococcus neoformans and Cryptococcus gattii genes preferentially expressed during rat macrophage infection. Med Mycol 2010, 48, 932–941. [Google Scholar] [CrossRef]
- Himmelreich, U.; Allen, C.; Dowd, S.; Malik, R.; Shehan, B.P.; Mountford, C.; Sorrell, T.C. Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes Infect 2003, 5, 285–290. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, P.Y.; Sham, A.; Perfect, J.R.; Kronstad, J.W. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol 2008, 69, 1456–1475. [Google Scholar] [CrossRef]
- Kronstad, J.; Saikia, S.; Nielson, E.D.; Kretschmer, M.; Jung, W.; Hu, G.; Geddes, J.M.; Griffiths, E.J.; Choi, J.; Cadieux, B.; et al. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell 2012, 11, 109–118. [Google Scholar] [CrossRef]
- Chun, C.D.; Brown, J.C.S.; Madhani, H.D. A major role for capsule-independent phagocytosis-inhibitory mechanisms in mammalian infection by Cryptococcus neoformans. Cell Host Microbe 2011, 9, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Chun, C.D.; Liu, O.W.; Madhani, H.D. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog 2007, 3, e22. [Google Scholar] [CrossRef]
- Chun, C.D.; Madhani, H.D. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; D'Souza, C.A.; Cox, G.M.; Heitman, J. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 2004, 3, 14–26. [Google Scholar] [CrossRef]
- Lengeler, K.B.; Wang, P.; Cox, G.M.; Perfect, J.R.; Heitman, J. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc Natl Acad Sci U S A 2000, 97, 14455–14460. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Wickes, B.L.; Miller, G.F.; Penoyer, L.A.; Kwon-Chung, K.J. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med 2000, 191, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Wickes, B.L.; Edman, U.; Edman, J.C. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol 1997, 26, 951–960. [Google Scholar] [CrossRef]
- Yue, C.; Cavallo, L.M.; Alspaugh, J.A.; Wang, P.; Cox, G.M.; Perfect, J.R.; Heitman, J. The STE12α Homolog Is Required for Haploid Filamentation But Largely Dispensable for Mating and Virulence in Cryptococcus neoformans. Genetics 1999, 153, 1601–1615. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Kojima, K.; Cox, G.M.; Heitman, J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell 2006, 17, 3122–3135. [Google Scholar] [CrossRef]
- Kojima, K.; Bahn, Y.S.; Heitman, J. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology (Reading) 2006, 152, 591–604. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Kojima, K.; Cox, G.M.; Heitman, J. Specialisation of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 2005, 16, 2285–2300. [Google Scholar] [CrossRef]
- Cruz, M.C.; Sia, R.A.L.; Olson, M.; Cox, G.M.; Heitman, J. Comparison of the Roles of Calcineurin in Physiology and Virulence in Serotype D and Serotype A Strains ofCryptococcus neoformans. Infection and Immunity 2000, 68, 982–985. [Google Scholar] [CrossRef]
- Yang, J.; Li, D.; Liu, X.; Pan, J.; Yan, B.; Zhu, X. Regulation of virulence factors, carbon utilisation and virulence by SNF1 in Cryptococcus neoformans JEC21 and divergent actions of SNF1 between cryptococcal strains. Fungal Genet Biol 2010, 47, 994–1000. [Google Scholar] [CrossRef]
- Free, S.J. Fungal cell wall organisation and biosynthesis. In Advances in genetics; Elsevier: 2013; Volume 81, pp. 33-82. [CrossRef]
- Lee, C.G.; Da Silva, C.A.; Dela Cruz, C.S.; Ahangari, F.; Ma, B.; Kang, M.J.; He, C.H.; Takyar, S.; Elias, J.A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 2011, 73, 479–501. [Google Scholar] [CrossRef]
- Fries, B.C.; Goldman, D.L.; Cherniak, R.; Ju, R.; Casadevall, A. Phenotypic Switching in Cryptococcus neoformans Results in Changes in Cellular Morphology and Glucuronoxylomannan Structure. Infection and immunity 1999, 67, 6076–6083. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Banks, I.R.; Specht, C.A.; Donlin, M.J.; Gerik, K.J.; Levitz, S.M.; Lodge, J.K. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 2005, 4, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Lodge, J.K. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot Cell 2011, 10, 1264–1268. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, R.; Lam, W.C.; Maybruck, B.; Specht, C.A.; Levitz, S.M.; Lodge, J.K. Induction of Protective Immunity to Cryptococcal Infection in Mice by a Heat-Killed, Chitosan-Deficient Strain of Cryptococcus neoformans. mBio 2016, 7, e00547–00516. [Google Scholar] [CrossRef] [PubMed]
- Ohtakara, A.; Izume, M.; Mitsutomi, M. Action of Microbial Chitinases on Chitosan with Different Degrees of Deacetylation. Agricultural and Biological Chemistry 2014, 52, 3181–3182. [Google Scholar] [CrossRef]
- Botts, M.R.; Hull, C.M. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol 2010, 13, 437–442. [Google Scholar] [CrossRef]
- Botts, M.R.; Giles, S.S.; Gates, M.A.; Kozel, T.R.; Hull, C.M. Isolation and characterisation of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell 2009, 8, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Sukroongreung, S.; Kitiniyom, K.; Nilakul, C.; Tantimavanich, S. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med Mycol 1998, 36, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Velagapudi, R.; Hsueh, Y.P.; Geunes-Boyer, S.; Wright, J.R.; Heitman, J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 2009, 77, 4345–4355. [Google Scholar] [CrossRef]
- Neilson, J.B.; Ivey, M.H.; Bulmer, G.S. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun 1978, 20, 262–266. [Google Scholar] [CrossRef]
- Lee, S.C.; Phadke, S.; Sun, S.; Heitman, J. Pseudohyphal growth of Cryptococcus neoformans is a reversible dimorphic transition in response to ammonium that requires Amt1 and Amt2 ammonium permeases. Eukaryot Cell 2012, 11, 1391–1398. [Google Scholar] [CrossRef]
- Wang, L.; Zhai, B.; Lin, X. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 2012, 8, e1002765. [Google Scholar] [CrossRef]
- Lin, X.; Jackson, J.C.; Feretzaki, M.; Xue, C.; Heitman, J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Magditch, D.A.; Liu, T.B.; Xue, C.; Idnurm, A. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog 2012, 8, e1002936. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Nielsen, K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 2012, 11, 820–826. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Wang, Y.; Ballou, E.R.; O'Meara, T.R.; Bahn, Y.S.; Alspaugh, J.A.; Xue, C.; Nielsen, K. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell 2011, 10, 1306–1316. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chretien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Alspaugh, J.A. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol 2015, 78, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Garcia-Rodas, R.; Nosanchuk, J.D.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L.; Casadevall, A. Fungal cell gigantism during mammalian infection. PLoS Pathog 2010, 6, e1000945. [Google Scholar] [CrossRef]
- Zaragoza, O.; Nielsen, K. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr Opin Microbiol 2013, 16, 409–413. [Google Scholar] [CrossRef]
- Idnurm, A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 2010, 185, 153–163. [Google Scholar] [CrossRef]
- Brandt, M.E.; Pfaller, M.A.; Hajjeh, R.A.; Graviss, E.A.; Rees, J.; Spitzer, E.D.; Pinner, R.W.; Mayer, L.W. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated Cryptococcosis. Cryptococcal Disease Active Surveillance Group. J Infect Dis 1996, 174, 812–820. [Google Scholar] [CrossRef]
- Fries, B.C.; Chen, F.; Currie, B.P.; Casadevall, A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol 1996, 34, 1531–1534. [Google Scholar] [CrossRef]
- Walton, F.J.; Heitman, J.; Idnurm, A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell 2006, 17, 3768–3780. [Google Scholar] [CrossRef] [PubMed]
- Kozubowski, L.; Aboobakar, E.F.; Cardenas, M.E.; Heitman, J. Calcineurin colocalises with P-bodies and stress granules during thermal stress in Cryptococcus neoformans. Eukaryot Cell 2011, 10, 1396–1402. [Google Scholar] [CrossRef]
- Nelson, B.; Kurischko, C.; Horecka, J.; Mody, M.; Nair, P.; Pratt, L.; Zougman, A.; McBroom, L.D.; Hughes, T.R.; Boone, C.; et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarised morphogenesis. Mol Biol Cell 2003, 14, 3782–3803. [Google Scholar] [CrossRef]
- Racki, W.J.; Becam, A.M.; Nasr, F.; Herbert, C.J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J 2000, 19, 4524–4532. [Google Scholar] [CrossRef]
- Zaragoza, O.; Rodrigues, M.L.; De Jesus, M.; Frases, S.; Dadachova, E.; Casadevall, A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 2009, 68, 133–216. [Google Scholar] [CrossRef]
- Vartivarian, S.E.; Anaissie, E.J.; Cowart, R.E.; Sprigg, H.A.; Tingler, M.J.; Jacobson, E.S. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis 1993, 167, 186–190. [Google Scholar] [CrossRef]
- Granger, D.L.; Perfect, J.R.; Durack, D.T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest 1985, 76, 508–516. [Google Scholar] [CrossRef]
- McFadden, D.C.; Fries, B.C.; Wang, F.; Casadevall, A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell 2007, 6, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Fries, B.C.; Cook, E.; Wang, X.; Casadevall, A. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob Agents Chemother 2005, 49, 350–357. [Google Scholar] [CrossRef]
- Vecchiarelli, A.; Retini, C.; Pietrella, D.; Monari, C.; Tascini, C.; Beccari, T.; Kozel, T.R. Downregulation by cryptococcal polysaccharide of TNFα and IL-1β secretion from human monocytes. Infect Immun 1995, 63, 2919–2923. [Google Scholar] [CrossRef] [PubMed]
- Vecchiarelli, A.; Retini, C.; Monari, C.; Tascini, C.; Bistoni, F.; Kozel, T.R. Purified capsular polysaccharide of Cryptococcus neoformans induces interleukin-10 secretion by human monocytes. Infect Immun 1996, 64, 2846–2849. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, E.S.; Compton, G.M. Discordant regulation of phenoloxidase and capsular polysaccharide in Cryptococcus neoformans. J Med Vet Mycol 1996, 34, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Reiss, E.; Cherniak, R.; Eby, R.; Kaufman, L. Enzyme immunoassay detection of IgM to galactoxylomannan of Cryptococcus neoformans. Diagn Immunol 1984, 2, 109–115. [Google Scholar] [PubMed]
- Dong, Z.M.; Murphy, J.W. Cryptococcal polysaccharides induce L-selectin shedding and tumor necrosis factor receptor loss from the surface of human neutrophils. J Clin Invest 1996, 97, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Ben-Abdallah, M.; Sturny-Leclere, A.; Ave, P.; Louise, A.; Moyrand, F.; Weih, F.; Janbon, G.; Memet, S. Fungal-induced cell cycle impairment, chromosome instability and apoptosis via differential activation of NF-kappaB. PLoS Pathog 2012, 8, e1002555. [Google Scholar] [CrossRef] [PubMed]
- Simonet, W.S.; Hughes, T.M.; Nguyen, H.Q.; Trebasky, L.D.; Danilenko, D.M.; Medlock, E.S. Long-term impaired neutrophil migration in mice overexpressing human interleukin-8. J Clin Invest 1994, 94, 1310–1319. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.K.; Bennett, J.E.; Glaudemans, C.P. Capsular polysaccharides of Cryptococcus neoformans. Rev Infect Dis 1984, 6, 619–624. [Google Scholar] [CrossRef]
- Fonseca, F.L.; Nohara, L.L.; Cordero, R.J.; Frases, S.; Casadevall, A.; Almeida, I.C.; Nimrichter, L.; Rodrigues, M.L. Immunomodulatory effects of serotype B glucuronoxylomannan from Cryptococcus gattii correlate with polysaccharide diameter. Infect Immun 2010, 78, 3861–3870. [Google Scholar] [CrossRef]
- Dong, Z.M.; Murphy, J.W. Effects of the two varieties of Cryptococcus neoformans cells and culture filtrate antigens on neutrophil locomotion. Infect Immun 1995, 63, 2632–2644. [Google Scholar] [CrossRef]
- Ko, Y.J.; Yu, Y.M.; Kim, G.B.; Lee, G.W.; Maeng, P.J.; Kim, S.; Floyd, A.; Heitman, J.; Bahn, Y.S. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 2009, 8, 1197–1217. [Google Scholar] [CrossRef]
- Lee, J.W.; Ko, Y.J.; Kim, S.Y.; Bahn, Y.S. Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Eukaryot Cell 2011, 10, 998–1002. [Google Scholar] [CrossRef]
- D'Souza, C.A.; Alspaugh, J.A.; Yue, C.; Harashima, T.; Cox, G.M.; Perfect, J.R.; Heitman, J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 2001, 21, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Alspaugh, J.A.; Perfect, J.R.; Heitman, J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 1997, 11, 3206–3217. [Google Scholar] [CrossRef]
- Cruickshank, J.G.; Cavill, R.; Jelbert, M. Cryptococcus neoformans of unusual morphology. Appl Microbiol 1973, 25, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Love, G.L.; Boyd, G.D.; Greer, D.L. Large Cryptococcus neoformans isolated from brain abscess. J Clin Microbiol 1985, 22, 1068–1070. [Google Scholar] [CrossRef]
- Li, L.; Shen, G.; Zhang, Z.G.; Wang, Y.L.; Thompson, J.K.; Wang, P. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell 2007, 18, 4201–4209. [Google Scholar] [CrossRef]
- Lian, T.; Simmer, M.I.; D'Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.J.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 2005, 55, 1452–1472. [Google Scholar] [CrossRef]
- Polacheck, I.; Hearing, V.J.; Kwon-Chung, K.J. Biochemical studies of phenoloxidase and utilisation of catecholamines in Cryptococcus neoformans. J Bacteriol 1982, 150, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Moyrand, F.; Chang, Y.C.; Himmelreich, U.; Kwon-Chung, K.J.; Janbon, G. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot Cell 2004, 3, 1513–1524. [Google Scholar] [CrossRef]
- Kim, M.S.; Ko, Y.J.; Maeng, S.; Floyd, A.; Heitman, J.; Bahn, Y.S. Comparative transcriptome analysis of the CO2 sensing pathway via differential expression of carbonic anhydrase in Cryptococcus neoformans. Genetics 2010, 185, 1207–1219. [Google Scholar] [CrossRef]
- Nurudeen, T.A.; Ahearn, D.G. Regulation of melanin production by Cryptococcus neoformans. J Clin Microbiol 1979, 10, 724–729. [Google Scholar] [CrossRef]
- Hamilton, A. Cryptococcus neoformans-the encapsulated menace. Mycologist 2002, 16, 125–126. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Rudolph, J.; Rosas, A.L.; Casadevall, A. Evidence that Cryptococcus neoformans is melanized in pigeon excreta: implications for pathogenesis. Infect Immun 1999, 67, 5477–5479. [Google Scholar] [CrossRef] [PubMed]
- Frases, S.; Salazar, A.; Dadachova, E.; Casadevall, A. Cryptococcus neoformans can utilise the bacterial melanin precursor homogentisic acid for fungal melanogenesis. Appl Environ Microbiol 2007, 73, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Frases, S.; Chaskes, S.; Dadachova, E.; Casadevall, A. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl Environ Microbiol 2006, 72, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Piattelli, M.; Fattorusso, E.; Magno, S.; Nicolaus, R.A. Ustilago Melanin, a Naturally Occurring Catechol Melanin. Tetrahedron Letters 1963, 4, 997–998. [Google Scholar] [CrossRef]
- Wang, Y.; Aisen, P.; Casadevall, A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun 1995, 63, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.E.; Wheeler, M.H.; Szaniszlo, P.J. Evidence for Pentaketide Melanin Biosynthesis in Dematiaceous Human Pathogenic Fungi. Mycologia 1987, 79, 320–322. [Google Scholar] [CrossRef]
- Stüssi, H.; Rast, D.M. The biosynthesis and possible function of γ-glutaminyl-4-hydroxybenzene in Agaricus bisporus. Phytochemistry 1981, 20, 2347–2352. [Google Scholar] [CrossRef]
- Leonowicz, A.; Cho, N.S.; Luterek, J.; Wilkolazka, A.; Wojtas-Wasilewska, M.; Matuszewska, A.; Hofrichter, M.; Wesenberg, D.; Rogalski, J. Fungal laccase: properties and activity on lignin. J Basic Microbiol 2001, 41, 185–227. [Google Scholar] [CrossRef]
- Garcia-Rivera, J.; Eisenman, H.C.; Nosanchuk, J.D.; Aisen, P.; Zaragoza, O.; Moadel, T.; Dadachova, E.; Casadevall, A. Comparative analysis of Cryptococcus neoformans acid-resistant particles generated from pigmented cells grown in different laccase substrates. Fungal Genet Biol 2005, 42, 989–998. [Google Scholar] [CrossRef]
- Chaskes, S.; Tyndall, R.L. Pigment production by Cryptococcus neoformans from para- and ortho-Diphenols: effect of the nitrogen source. J Clin Microbiol 1975, 1, 509–514. [Google Scholar] [CrossRef]
- Wheeler, M.H.; Bell, A.A. Melanins and their importance in pathogenic fungi. In Current topics in medical mycology; Springer: 1988; pp. 338-387. [CrossRef]
- Maeda, T. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J 2012, 279, 1407–1413. [Google Scholar] [CrossRef]
- Godinho, R.M.; Crestani, J.; Kmetzsch, L.; Araujo Gde, S.; Frases, S.; Staats, C.C.; Schrank, A.; Vainstein, M.H.; Rodrigues, M.L. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep 2014, 4, 6198. [Google Scholar] [CrossRef] [PubMed]
- Sussman, A.S. The Fungal Population: An Advanced Treatise; Elsevier Science: 2013; pp. 447-486.
- Garcia-Rivera, J.; Casadevall, A. Melanisation of Cryptococcus neoformans reduces its susceptibility to the antimicrobial effects of silver nitrate. Med Mycol 2001, 39, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Casadevall, A. Growth of Cryptococcus neoformans in presence of L-dopa decreases its susceptibility to amphotericin B. Antimicrob Agents Chemother 1994, 38, 2648–2650. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Susceptibility of melanised and nonmelanised Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun 1994, 62, 3004–3007. [Google Scholar] [CrossRef] [PubMed]
- Durrell, L.W. The Composition and Structure of Walls of Dark Fungus Spores. Mycopathol Mycol Appl 1964, 23, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Rosas, A.L.; Casadevall, A. Melanisation affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol Lett 1997, 153, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Morison, W.L. What is the function of melanin? Archives of dermatology 1985, 121, 1160–1163. [Google Scholar] [CrossRef]
- Hill, H.Z. The function of melanin or six blind people examine an elephant. Bioessays 1992, 14, 49–56. [Google Scholar] [CrossRef]
- Wang, Y.; Aisen, P.; Casadevall, A. Melanin, melanin "ghosts," and melanin composition in Cryptococcus neoformans. Infect Immun 1996, 64, 2420–2424. [Google Scholar] [CrossRef]
- Rosas, A.L.; Nosanchuk, J.D.; Gomez, B.L.; Edens, W.A.; Henson, J.M.; Casadevall, A. Isolation and serological analyses of fungal melanins. J Immunol Methods 2000, 244, 69–80. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Susceptibility of melanised and nonmelanised Cryptococcus neoformans to the melanin-binding compounds trifluoperazine and chloroquine. Antimicrob Agents Chemother 1996, 40, 541–545. [Google Scholar] [CrossRef]
- Doering, T.L.; Nosanchuk, J.D.; Roberts, W.K.; Casadevall, A. Melanin as a potential cryptococcal defence against microbicidal proteins. Medical Mycology 1999, 37, 175–181. [Google Scholar] [CrossRef]
- Zhu, X.; Gibbons, J.; Zhang, S.; Williamson, P.R. Copper-mediated reversal of defective laccase in a Deltavph1 avirulent mutant of Cryptococcus neoformans. Mol Microbiol 2003, 47, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Siafakas, A.R.; Wright, L.C.; Sorrell, T.C.; Djordjevic, J.T. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell 2006, 5, 488–498. [Google Scholar] [CrossRef]
- Williamson, P.R. Biochemical and molecular characterisation of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol 1994, 176, 656–664. [Google Scholar] [CrossRef]
- Torres-Guererro, H.; Edman, J.C. Melanin-deficient mutants of Cryptococcus neoformans. J Med Vet Mycol 1994, 32, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Gerrald, Q.D.; Kraus, P.R.; Boily, M.J.; Davis, M.J.; Giles, S.S.; Cox, G.M.; Heitman, J.; Alspaugh, J.A. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell 2005, 4, 190–201. [Google Scholar] [CrossRef]
- Lee, D.; Jang, E.H.; Lee, M.; Kim, S.W.; Lee, Y.; Lee, K.T.; Bahn, Y.S. Unraveling Melanin Biosynthesis and Signaling Networks in Cryptococcus neoformans. mBio 2019, 10, e02267–02219. [Google Scholar] [CrossRef] [PubMed]
- Walton, F.J.; Idnurm, A.; Heitman, J. Novel gene functions required for melanisation of the human pathogen Cryptococcus neoformans. Mol Microbiol 2005, 57, 1381–1396. [Google Scholar] [CrossRef]
- Song, M.H.; Lee, J.W.; Kim, M.S.; Yoon, J.K.; White, T.C.; Floyd, A.; Heitman, J.; Strain, A.K.; Nielsen, J.N.; Nielsen, K.; et al. A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryot Cell 2012, 11, 53–67. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Rosas, A.L.; Lee, S.C.; Casadevall, A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 2000, 355, 2049–2050. [Google Scholar] [CrossRef]
- Rosas, A.L.; Nosanchuk, J.D.; Feldmesser, M.; Cox, G.M.; McDade, H.C.; Casadevall, A. Synthesis of polymerised melanin by Cryptococcus neoformans in infected rodents. Infect Immun 2000, 68, 2845–2853. [Google Scholar] [CrossRef]
- Liu, L.; Wakamatsu, K.; Ito, S.; Williamson, P.R. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect Immun 1999, 67, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Wakamatsu, K. Chemical degradation of melanins: application to identification of dopamine-melanin. Pigment Cell Res 1998, 11, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.; Pezzella, A.; d'Ischia, M.; Prota, G. New pyrrole acids by oxidative degradation of eumelanins with hydrogen peroxide. Further hints to the mechanism of pigment breakdown. Tetrahedron 1996, 52, 8775–8780. [Google Scholar] [CrossRef]
- Napolitano, A.; Pezzella, A.; Vincensi, M.R.; Prota, G. Oxidative degradation of melanins to pyrrole acids: A model study. Tetrahedron 1995, 51, 5913–5920. [Google Scholar] [CrossRef]
- Wang, P.; Cutler, J.; King, J.; Palmer, D. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot Cell 2004, 3, 1028–1035. [Google Scholar] [CrossRef]
- Luberto, C.; Toffaletti, D.L.; Wills, E.A.; Tucker, S.C.; Casadevall, A.; Perfect, J.R.; Hannun, Y.A.; Del Poeta, M. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev 2001, 15, 201–212. [Google Scholar] [CrossRef]
- Alspaugh, J.A.; Cavallo, L.M.; Perfect, J.R.; Heitman, J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol 2000, 36, 352–365. [Google Scholar] [CrossRef]
- Waugh, M.S.; Nichols, C.B.; DeCesare, C.M.; Cox, G.M.; Heitman, J.; Alspaugh, J.A. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology (Reading) 2002, 148, 191–201. [Google Scholar] [CrossRef]
- Maeng, S.; Ko, Y.J.; Kim, G.B.; Jung, K.W.; Floyd, A.; Heitman, J.; Bahn, Y.S. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell 2010, 9, 360–378. [Google Scholar] [CrossRef]
- Liu, K.H.; Shen, W.C. Mating differentiation in Cryptococcus neoformans is negatively regulated by the Crk1 protein kinase. Fungal Genet Biol 2011, 48, 225–240. [Google Scholar] [CrossRef]
- Wang, P.; Heitman, J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol 1999, 2, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Perfect, J.R.; Heitman, J. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Molecular and Cellular Biology 2000, 20, 352–362. [Google Scholar] [CrossRef]
- Clarke, D.L.; Woodlee, G.L.; McClelland, C.M.; Seymour, T.S.; Wickes, B.L. The Cryptococcus neoformans STE11alpha gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol Microbiol 2001, 40, 200–213. [Google Scholar] [CrossRef]
- Wang, P.; Nichols, C.B.; Lengeler, K.B.; Cardenas, M.E.; Cox, G.M.; Perfect, J.R.; Heitman, J. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot Cell 2002, 1, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Perfect, J.R.; Heitman, J. The G-protein β subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Molecular and Cellular Biology 2000, 20, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Leeuw, T.; Wu, C.; Schrag, J.D.; Whiteway, M.; Thomas, D.Y.; Leberer, E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 1998, 391, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Kim, S.Y.; Okagaki, L.H.; Nielsen, K.; Bahn, Y.S. Ste50 adaptor protein governs sexual differentiation of Cryptococcus neoformans via the pheromone-response MAPK signaling pathway. Fungal Genet Biol 2011, 48, 154–165. [Google Scholar] [CrossRef]
- Lengeler, K.B.; Cox, G.M.; Heitman, J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun 2001, 69, 115–122. [Google Scholar] [CrossRef]
- Kraus, P.R.; Heitman, J. Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem Biophys Res Commun 2003, 311, 1151–1157. [Google Scholar] [CrossRef]
- Ma, P.; Wera, S.; Van Dijck, P.; Thevelein, J.M. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol Biol Cell 1999, 10, 91–104. [Google Scholar] [CrossRef]
- Maliehe, M.; Ntoi, M.A.; Lahiri, S.; Folorunso, O.S.; Ogundeji, A.O.; Pohl, C.H.; Sebolai, O.M. Environmental Factors That Contribute to the Maintenance of Cryptococcus neoformans Pathogenesis. Microorganisms 2020, 8, 180. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Cox, G.M.; Perfect, J.R.; Heitman, J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol 2005, 15, 2013–2020. [Google Scholar] [CrossRef]
- Klengel, T.; Liang, W.J.; Chaloupka, J.; Ruoff, C.; Schroppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 2005, 15, 2021–2026. [Google Scholar] [CrossRef]
- Gyawali, R.; Zhao, Y.; Lin, J.; Fan, Y.; Xu, X.; Upadhyay, S.; Lin, X. Pheromone independent unisexual development in Cryptococcus neoformans. PLoS Genet 2017, 13, e1006772. [Google Scholar] [CrossRef]
- Feretzaki, M.; Billmyre, R.B.; Clancey, S.A.; Wang, X.; Heitman, J. Gene Network Polymorphism Illuminates Loss and Retention of Novel RNAi Silencing Components in the Cryptococcus Pathogenic Species Complex. PLoS Genet 2016, 12, e1005868. [Google Scholar] [CrossRef]
- Catalanotto, C.; Pallotta, M.; ReFalo, P.; Sachs, M.S.; Vayssie, L.; Macino, G.; Cogoni, C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol 2004, 24, 2536–2545. [Google Scholar] [CrossRef]
- Maiti, M.; Lee, H.C.; Liu, Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev 2007, 21, 590–600. [Google Scholar] [CrossRef]
- Catalanotto, C.; Azzalin, G.; Macino, G.; Cogoni, C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev 2002, 16, 790–795. [Google Scholar] [CrossRef]
- Jacobson, E.S.; Tingler, M.J.; Quynn, P.L. Effect of Hypertonic Solutes Upon the Polysaccharide Capsule in Cryptococcus neoformans: Die Wirkung hypertonischer Lösungen auf die Polysaccharid-Kapsel von Cryptococcus neofomans. Mycoses 1989, 32, 14–23. [Google Scholar] [CrossRef]
- Bahn, Y.S. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell 2008, 7, 2017–2036. [Google Scholar] [CrossRef]
- Nichols, C.B.; Perfect, Z.H.; Alspaugh, J.A. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 2007, 63, 1118–1130. [Google Scholar] [CrossRef]
- Chang, Y.C.; Penoyer, L.A. Properties of various Rho1 mutant alleles of Cryptococcus neoformans. J Bacteriol 2000, 182, 4987–4991. [Google Scholar] [CrossRef]
- Nonaka, H.; Tanaka, K.; Hirano, H.; Fujiwara, T.; Kohno, H.; Umikawa, M.; Mino, A.; Takai, Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. The EMBO journal 1995, 14, 5931–5938. [Google Scholar] [CrossRef]
- Lam, W.C.; Gerik, K.J.; Lodge, J.K. Role of Cryptococcus neoformans Rho1 GTPases in the PKC1 signaling pathway in response to thermal stress. Eukaryot Cell 2013, 12, 118–131. [Google Scholar] [CrossRef]
- Gerik, K.J.; Bhimireddy, S.R.; Ryerse, J.S.; Specht, C.A.; Lodge, J.K. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 2008, 7, 1685–1698. [Google Scholar] [CrossRef]
- Gerik, K.J.; Donlin, M.J.; Soto, C.E.; Banks, A.M.; Banks, I.R.; Maligie, M.A.; Selitrennikoff, C.P.; Lodge, J.K. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol Microbiol 2005, 58, 393–408. [Google Scholar] [CrossRef]
- Hu, G.; Steen, B.R.; Lian, T.; Sham, A.P.; Tam, N.; Tangen, K.L.; Kronstad, J.W. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog 2007, 3, e42. [Google Scholar] [CrossRef]
- Goebl, M.; Yanagida, M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends in biochemical sciences 1991, 16, 173–177. [Google Scholar] [CrossRef]
- Lamb, J.R.; Tugendreich, S.; Hieter, P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends in biochemical sciences 1995, 20, 257–259. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Michaud, W.A.; Wootton, J.C.; Boguski, M.S.; Connelly, C.; Hieter, P. TPR proteins as essential components of the yeast cell cycle. 1991, 1991; pp. 663-673.
- Gindhart, J.G., Jr.; Goldstein, L.S. Tetratrico peptide repeats are present in the kinesin light chain. Trends in biochemical sciences 1996, 21, 52–53. [Google Scholar] [CrossRef]
- Kyrpides, N.C.; Woese, C.R. Tetratrico-peptide-repeat proteins in the archaeon Methanococcus jannaschii. Trends in biochemical sciences 1998, 23, 245–247. [Google Scholar] [CrossRef]
- Zhu, W.; Rainville, I.R.; Ding, M.; Bolus, M.; Heintz, N.H.; Pederson, D.S. Evidence that the pre-mRNA splicing factor Clf1p plays a role in DNA replication in Saccharomyces cerevisiae. Genetics 2002, 160, 1319–1333. [Google Scholar] [CrossRef]
- Zhang, K.; Smouse, D.; Perrimon, N. The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes & Development 1991, 5, 1080–1091. [Google Scholar]
- Chung, S.; McLean, M.R.; Rymond, B.C. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA (New York, N.Y.) 1999, 5, 1042–1054. [Google Scholar] [CrossRef]
- Russell, C.S.; Ben-Yehuda, S.; Dix, I.; Kupiec, M.; Beggs, J.D. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA (New York, N.Y.) 2000, 6, 1565–1572. [Google Scholar] [CrossRef]
- Chung, S.; Mondon, P.; Chang, Y.C.; Kwon-Chung, K.J. Cryptococcus neoformans with a mutation in the tetratricopeptide repeat-containing gene, CCN1, causes subcutaneous lesions but fails to cause systemic infection. Infect Immun 2003, 71, 1988–1994. [Google Scholar] [CrossRef]
- Bemis, D.A.; Krahwinkel, D.J.; Bowman, L.A.; Mondon, P.; Kwon-Chung, K.J. Temperature-sensitive strain of Cryptococcus neoformans producing hyphal elements in a feline nasal granuloma. J Clin Microbiol 2000, 38, 926–928. [Google Scholar] [CrossRef]
- Park, H.S.; Chow, E.W.; Fu, C.; Soderblom, E.J.; Moseley, M.A.; Heitman, J.; Cardenas, M.E. Calcineurin Targets Involved in Stress Survival and Fungal Virulence. PLoS Pathog 2016, 12, e1005873. [Google Scholar] [CrossRef]
- Kraus, P.R.; Boily, M.J.; Giles, S.S.; Stajich, J.E.; Allen, A.; Cox, G.M.; Dietrich, F.S.; Perfect, J.R.; Heitman, J. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot Cell 2004, 3, 1249–1260. [Google Scholar] [CrossRef]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.C.; Cavallo, L.M.; Gorlach, J.M.; Cox, G.; Perfect, J.R.; Cardenas, M.E.; Heitman, J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol 1999, 19, 4101–4112. [Google Scholar] [CrossRef] [PubMed]
- Odom, A.; Del Poeta, M.; Perfect, J.; Heitman, J. The immunosuppressant FK506 and its nonimmunosuppressive analog L-685,818 are toxic to Cryptococcus neoformans by inhibition of a common target protein. Antimicrob Agents Chemother 1997, 41, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Fischer, G. Peptidyl-prolyl cis/trans isomerases and their effectors. Angewandte Chemie International Edition in English 1994, 33, 1415–1436. [Google Scholar] [CrossRef]
- Fischer, G.; Tradler, T.; Zarnt, T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett 1998, 426, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Cardenas, M.E.; Cox, G.M.; Perfect, J.R.; Heitman, J. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep 2001, 2, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Rossettini, A.; Chaturvedi, V.; Hanes, S.D. The Ess1 prolyl isomerase is dispensable for growth but required for virulence in Cryptococcus neoformans. Microbiology (Reading) 2005, 151, 1593–1605. [Google Scholar] [CrossRef]
- Del Poeta, M.; Toffaletti, D.L.; Rude, T.H.; Dykstra, C.C.; Heitman, J.; Perfect, J.R. Topoisomerase I Is Essential in Cryptococcus neoformans: Role in Pathobiology and as an Antifungal Target. Genetics 1999, 152, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Wang, J.C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci U S A 1985, 82, 7178–7182. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.; Redinbo, M.R.; Qiu, X.; Hol, W.G.; Champoux, J.J. A model for the mechanism of human topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Gerhold, D.; Kmiec, E.B.; Hauser, M.; Becker, J.M.; Koltin, Y. The topoisomerase I gene from Candida albicans. Microbiology (Reading) 1997, 143 ( Pt 2) Pt 2, 377–386. [Google Scholar] [CrossRef]
- Shen, L.L.; Baranowski, J.; Fostel, J.; Montgomery, D.A.; Lartey, P.A. DNA topoisomerases from pathogenic fungi: targets for the discovery of antifungal drugs. Antimicrob Agents Chemother 1992, 36, 2778–2784. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Heitman, J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 2005, 3, e95. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Walton, F.J.; Floyd, A.; Reedy, J.L.; Heitman, J. Identification of ENA1 as a virulence gene of the human pathogenic fungus Cryptococcus neoformans through signature-tagged insertional mutagenesis. Eukaryot Cell 2009, 8, 315–326. [Google Scholar] [CrossRef]
- Jung, K.W.; Strain, A.K.; Nielsen, K.; Jung, K.H.; Bahn, Y.S. Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet Biol 2012, 49, 332–345. [Google Scholar] [CrossRef]
- Chayakulkeeree, M.; Sorrell, T.C.; Siafakas, A.R.; Wilson, C.F.; Pantarat, N.; Gerik, K.J.; Boadle, R.; Djordjevic, J.T. Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol Microbiol 2008, 69, 809–826. [Google Scholar] [CrossRef]
- Lev, S.; Desmarini, D.; Li, C.; Chayakulkeeree, M.; Traven, A.; Sorrell, T.C.; Djordjevic, J.T. Phospholipase C of Cryptococcus neoformans regulates homeostasis and virulence by providing inositol trisphosphate as a substrate for Arg1 kinase. Infect Immun 2013, 81, 1245–1255. [Google Scholar] [CrossRef]
- Prasad, T.; Chandra, A.; Mukhopadhyay, C.K.; Prasad, R. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother 2006, 50, 3597–3606. [Google Scholar] [CrossRef]
- Jung, K.W.; Yang, D.H.; Maeng, S.; Lee, K.T.; So, Y.S.; Hong, J.; Choi, J.; Byun, H.J.; Kim, H.; Bang, S.; et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun 2015, 6, 6757. [Google Scholar] [CrossRef]
- Chang, Y.C.; Bien, C.M.; Lee, H.; Espenshade, P.J.; Kwon-Chung, K.J. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol 2007, 64, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Kraus, P.R.; Fox, D.S.; Cox, G.M.; Heitman, J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol Microbiol 2003, 48, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Douglas, C.M.; Li, W.; Jue, C.K.; Pramanik, B.; Yuan, X.; Rude, T.H.; Toffaletti, D.L.; Perfect, J.R.; Kurtz, M. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol 1999, 181, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Wormley, F.L., Jr.; Heinrich, G.; Miller, J.L.; Perfect, J.R.; Cox, G.M. Identification and characterisation of an SKN7 homologue in Cryptococcus neoformans. Infect Immun 2005, 73, 5022–5030. [Google Scholar] [CrossRef]
- Coenjaerts, F.E.; Hoepelman, A.I.; Scharringa, J.; Aarts, M.; Ellerbroek, P.M.; Bevaart, L.; Van Strijp, J.A.; Janbon, G. The Skn7 response regulator of Cryptococcus neoformans is involved in oxidative stress signalling and augments intracellular survival in endothelium. FEMS Yeast Res 2006, 6, 652–661. [Google Scholar] [CrossRef]
- Missall, T.A.; Lodge, J.K. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol Microbiol 2005, 57, 847–858. [Google Scholar] [CrossRef]
- Missall, T.A.; Pusateri, M.E.; Lodge, J.K. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol Microbiol 2004, 51, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Missall, T.A.; Cherry-Harris, J.F.; Lodge, J.K. Two glutathione peroxidases in the fungal pathogen Cryptococcus neoformans are expressed in the presence of specific substrates. Microbiology (Reading) 2005, 151, 2573–2581. [Google Scholar] [CrossRef]
- Giles, S.S.; Batinic-Haberle, I.; Perfect, J.R.; Cox, G.M. Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot Cell 2005, 4, 46–54. [Google Scholar] [CrossRef]
- Narasipura, S.D.; Chaturvedi, V.; Chaturvedi, S. Characterisation of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol Microbiol 2005, 55, 1782–1800. [Google Scholar] [CrossRef]
- Upadhya, R.; Campbell, L.T.; Donlin, M.J.; Aurora, R.; Lodge, J.K. Global transcriptome profile of Cryptococcus neoformans during exposure to hydrogen peroxide induced oxidative stress. PLoS One 2013, 8, e55110. [Google Scholar] [CrossRef] [PubMed]
- Demasi, A.P.; Pereira, G.A.; Netto, L.E. Yeast oxidative stress response. Influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. FEBS J 2006, 273, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.H.; Seyfang, A. High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology (Reading) 2003, 149, 3371–3381. [Google Scholar] [CrossRef]
- Crabtree, R.H. The organometallic chemistry of the transition metals; John Wiley & Sons: 2009.
- Giles, S.S.; Stajich, J.E.; Nichols, C.; Gerrald, Q.D.; Alspaugh, J.A.; Dietrich, F.; Perfect, J.R. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell 2006, 5, 1447–1459. [Google Scholar] [CrossRef]
- Lu, J.M.; Deschenes, R.J.; Fassler, J.S. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot Cell 2003, 2, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.B.; Ferreyra, J.; Ballou, E.R.; Alspaugh, J.A. Subcellular localisation directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot Cell 2009, 8, 181–189. [Google Scholar] [CrossRef]
- Sato, N.; Kawahara, H.; Toh-e, A.; Maeda, T. Phosphorelay-regulated degradation of the yeast Ssk1p response regulator by the ubiquitin-proteasome system. Mol Cell Biol 2003, 23, 6662–6671. [Google Scholar] [CrossRef]
- Cheng, L.; Watt, R.; Piper, P.W. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol Gen Genet 1994, 243, 358–362. [Google Scholar] [CrossRef]
- Walsh, L.; Schmuckli-Maurer, J.; Billinton, N.; Barker, M.G.; Heyer, W.D.; Walmsley, R.M. DNA-damage induction of RAD54 can be regulated independently of the RAD9- and DDC1-dependent checkpoints that regulate RNR2. Curr Genet 2002, 41, 232–240. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; La Sorda, M.; Torelli, R.; Fiori, B.; Santangelo, R.; Delogu, G.; Fadda, G. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect Immun 2006, 74, 1352–1359. [Google Scholar] [CrossRef]
- Lee, H.; Chang, Y.C.; Nardone, G.; Kwon-Chung, K.J. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol Microbiol 2007, 64, 591–601. [Google Scholar] [CrossRef]
- Tian, X.; He, G.J.; Hu, P.; Chen, L.; Tao, C.; Cui, Y.L.; Shen, L.; Ke, W.; Xu, H.; Zhao, Y.; et al. Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nat Microbiol 2018, 3, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Homer, C.M.; Summers, D.K.; Goranov, A.I.; Clarke, S.C.; Wiesner, D.L.; Diedrich, J.K.; Moresco, J.J.; Toffaletti, D.; Upadhya, R.; Caradonna, I.; et al. Intracellular Action of a Secreted Peptide Required for Fungal Virulence. Cell Host Microbe 2016, 19, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.A.; Jung, K.W.; Chen, Y.L.; Heitman, J.; Bahn, Y.S.; Kang, H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog 2011, 7, e1002177. [Google Scholar] [CrossRef] [PubMed]
- Glazier, V.E.; Kaur, J.N.; Brown, N.T.; Rivera, A.A.; Panepinto, J.C. Puf4 regulates both splicing and decay of HXL1 mRNA encoding the unfolded protein response transcription factor in Cryptococcus neoformans. Eukaryot Cell 2015, 14, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Havel, V.E.; Wool, N.K.; Ayad, D.; Downey, K.M.; Wilson, C.F.; Larsen, P.; Djordjevic, J.T.; Panepinto, J.C. Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans. Eukaryot Cell 2011, 10, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Kang, H.A.; Bahn, Y.S. Essential roles of the Kar2/BiP molecular chaperone downstream of the UPR pathway in Cryptococcus neoformans. PLoS One 2013, 8, e58956. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 2000, 279, 445–450. [Google Scholar] [CrossRef]
- Pincus, D.; Chevalier, M.W.; Aragon, T.; van Anken, E.; Vidal, S.E.; El-Samad, H.; Walter, P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 2010, 8, e1000415. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Vishwanath, P.; Cui, J.; Kelleher, D.J.; Gilmore, R.; Robbins, P.W.; Samuelson, J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci U S A 2007, 104, 11676–11681. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.T.; Byun, H.J.; Jung, K.W.; Hong, J.; Cheong, E.; Bahn, Y.S. Distinct and redundant roles of protein tyrosine phosphatases Ptp1 and Ptp2 in governing the differentiation and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 2014, 13, 796–812. [Google Scholar] [CrossRef]
- Erickson, T.; Liu, L.; Gueyikian, A.; Zhu, X.; Gibbons, J.; Williamson, P.R. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol Microbiol 2001, 42, 1121–1131. [Google Scholar] [CrossRef]
- Kiewietdejonge, A.; Pitts, M.; Cabuhat, L.; Sherman, C.; Kladwang, W.; Miramontes, G.; Floresvillar, J.; Chan, J.; Ramirez, R.M. Hypersaline stress induces the turnover of phosphatidylcholine and results in the synthesis of the renal osmoprotectant glycerophosphocholine in Saccharomyces cerevisiae. FEMS Yeast Res 2006, 6, 205–217. [Google Scholar] [CrossRef]
- Zablocki, K.; Miller, S.P.; Garcia-Perez, A.; Burg, M.B. Accumulation of glycerophosphocholine (GPC) by renal cells: osmotic regulation of GPC:choline phosphodiesterase. Proc Natl Acad Sci U S A 1991, 88, 7820–7824. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




