Submitted:
06 July 2023
Posted:
07 July 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Screening for bacteriophages:
2.2. Isolation and purification of bacteriophages:
2.3. TEM imaging:
2.4. One step growth curve:
2.5. Host specificity:
2.6. Temperature and pH stability:
2.7. In vitro time kill study:
2.8. In vivo toxicity study:
2.9. In vivo infection:
2.10. Genome sequence and analysis:
2.11. E. coli bacteriophage host receptor prediction using Machine Learning based multi class classifier:
3. Results
3.1. Identification of phage against XDR E. coli strains:
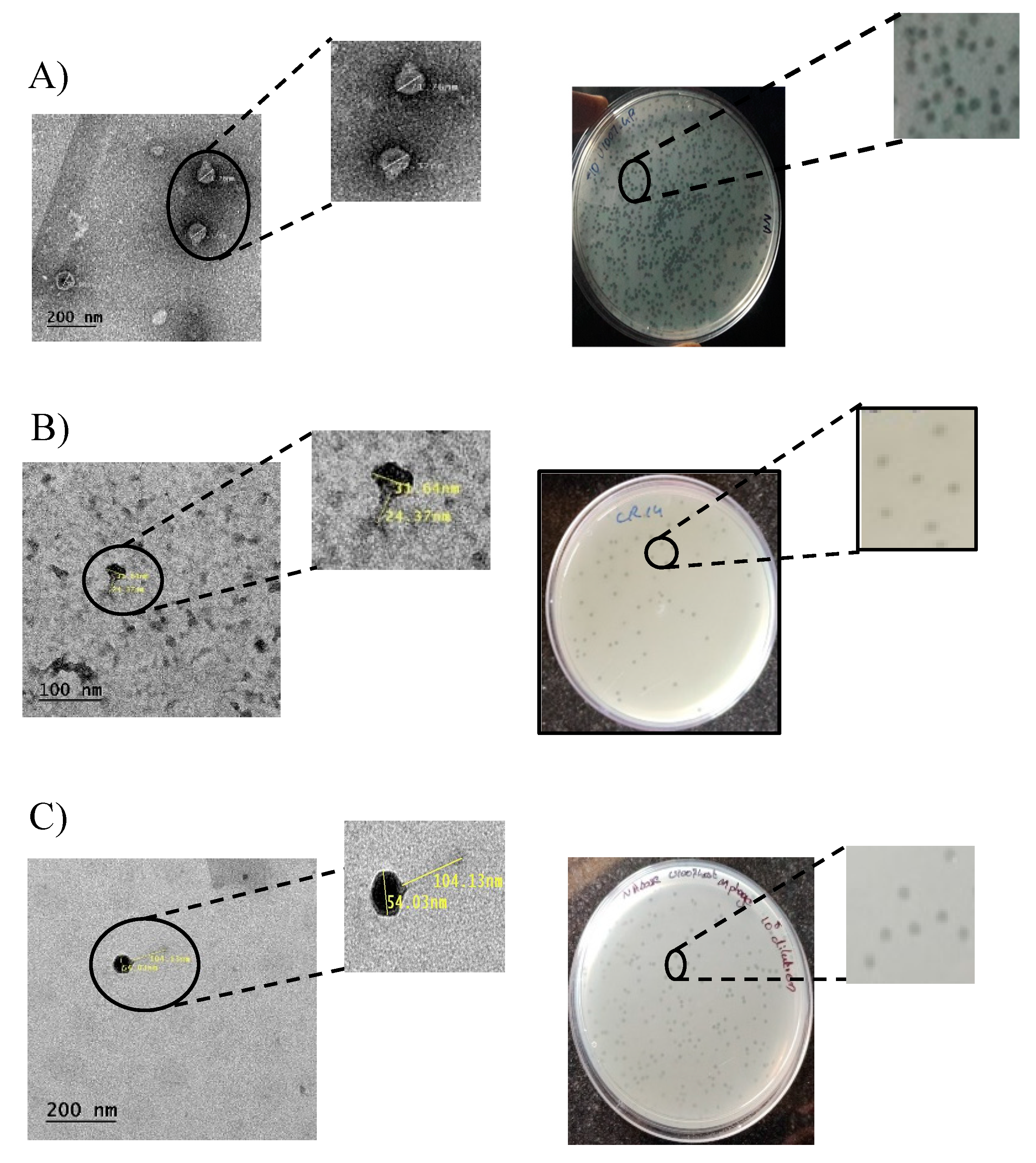
3.2. Isolation and purification of U1G, CR and M phages:
3.3. TEM imaging:
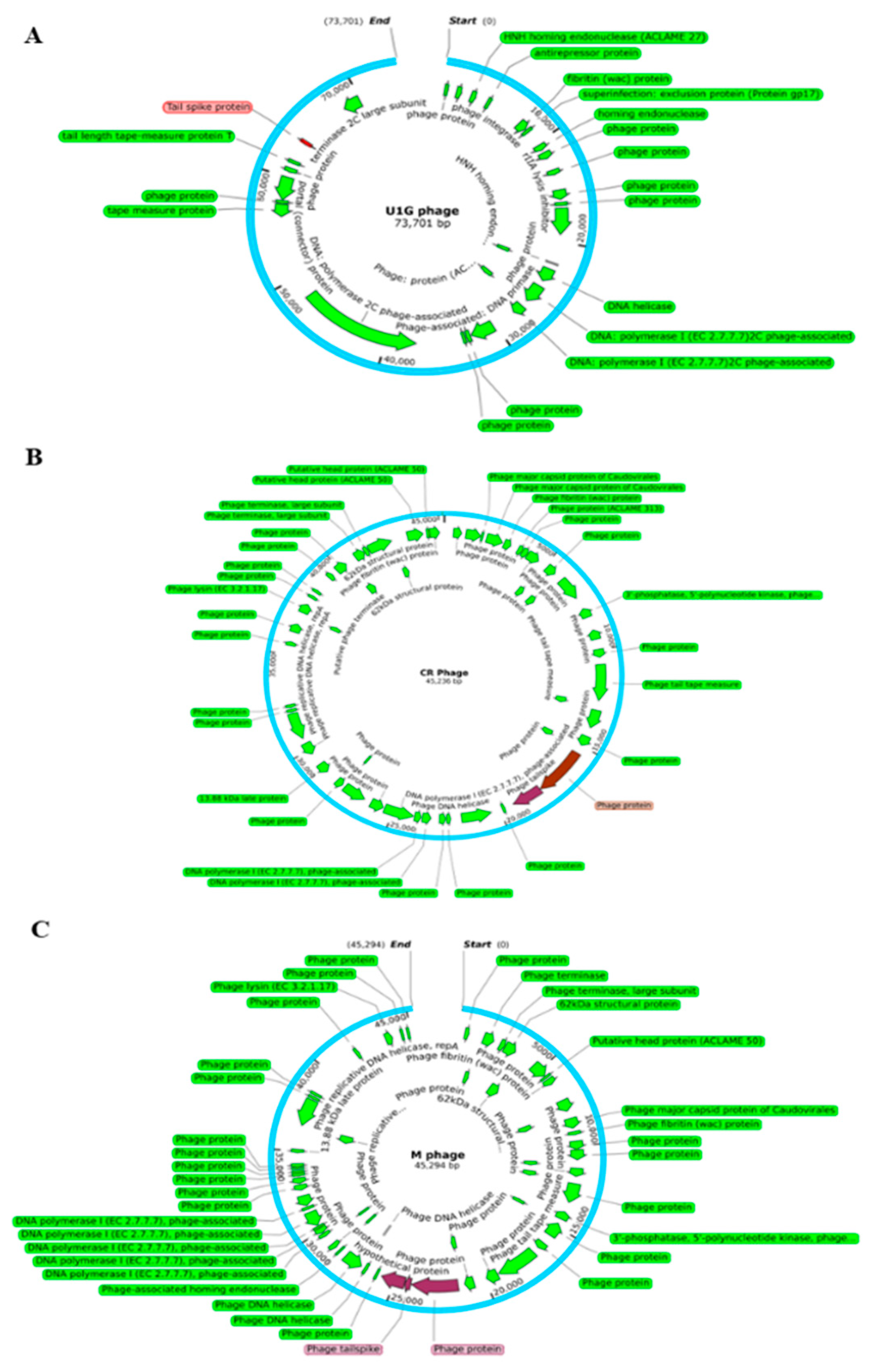
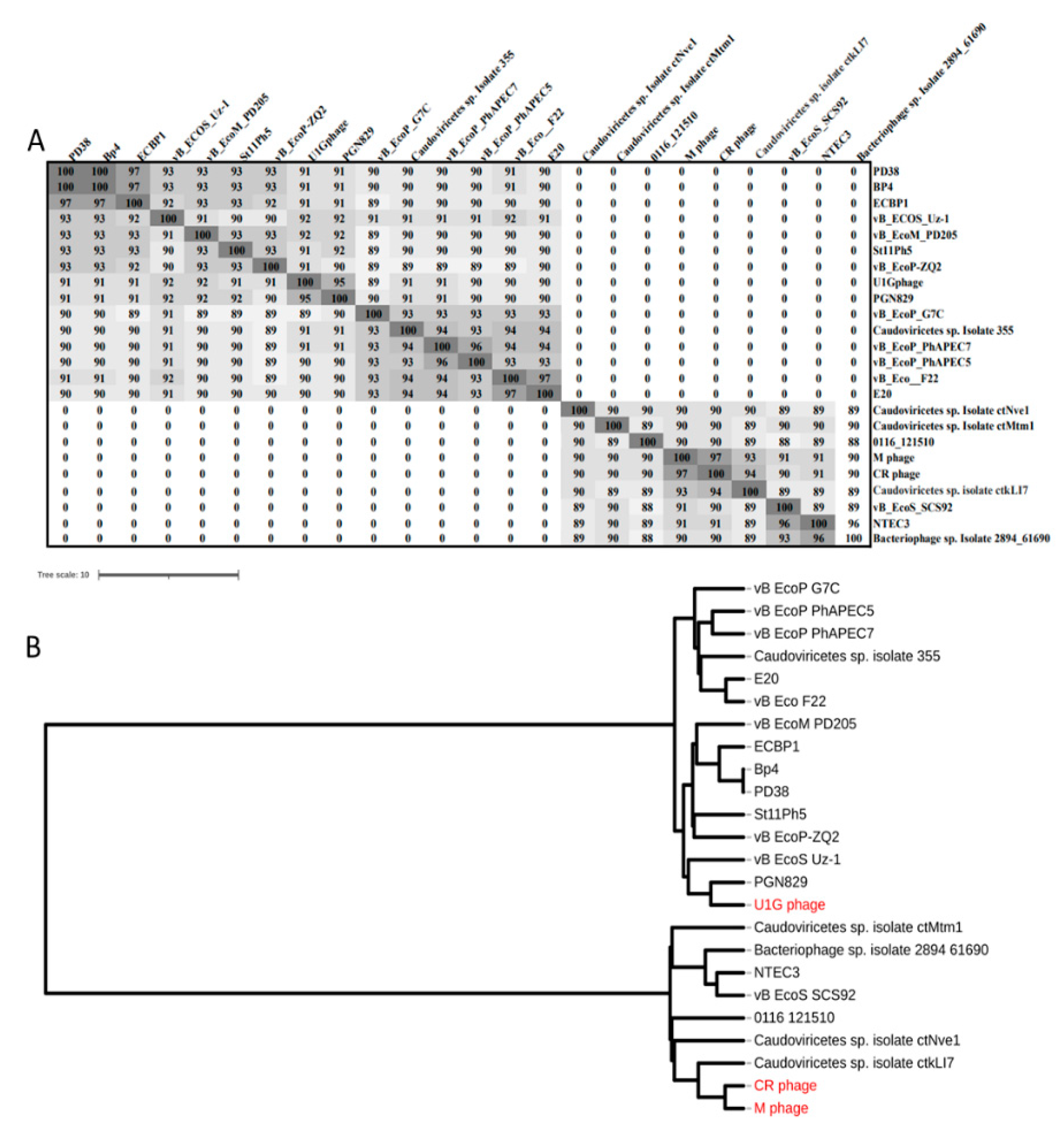
3.4. Genome analysis:
3.5. A multiclass-classification model for bacteriophage host-receptor prediction in E. coli:
Physiological Validation of host OmpC as the receptor for U1G phage
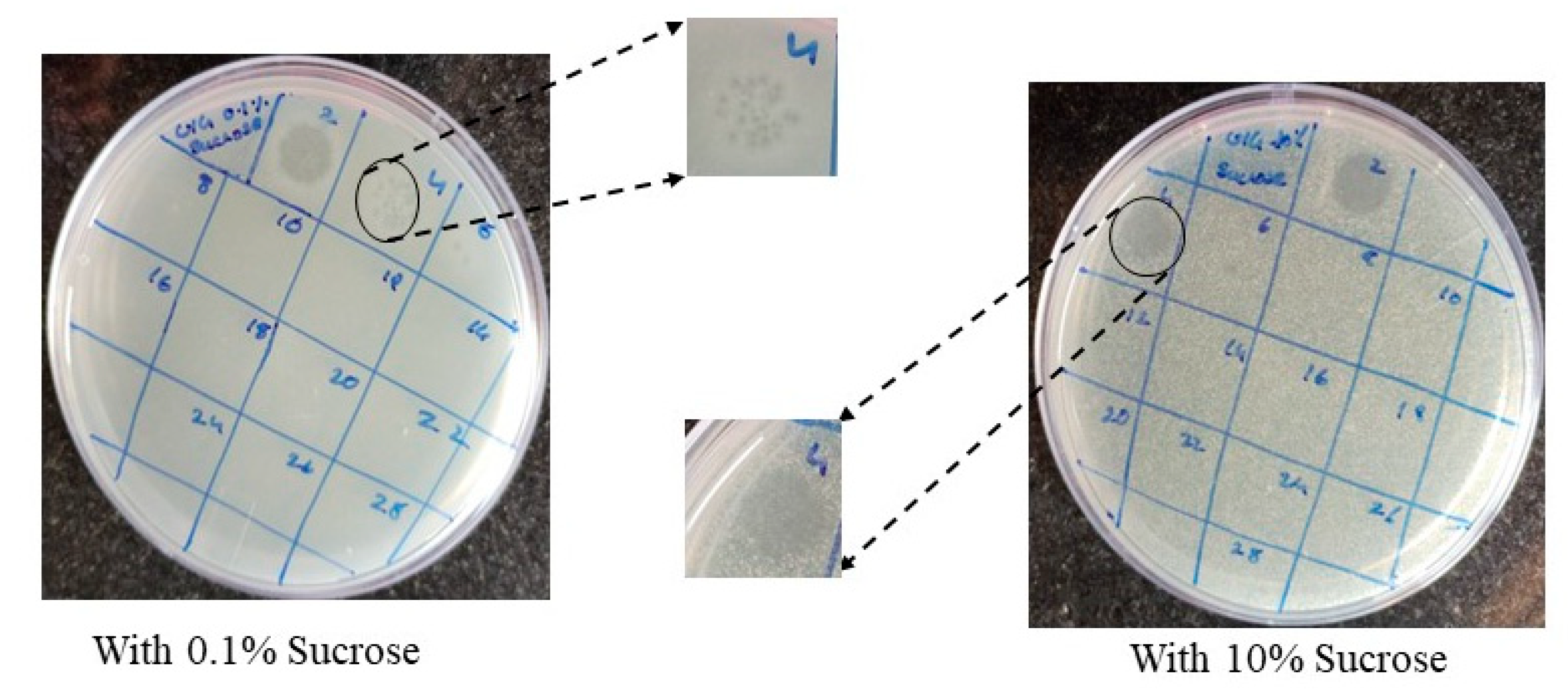
3.6. Burst size, Latent Period and Host Specificity:
3.7. pH and temperature sensitivity:
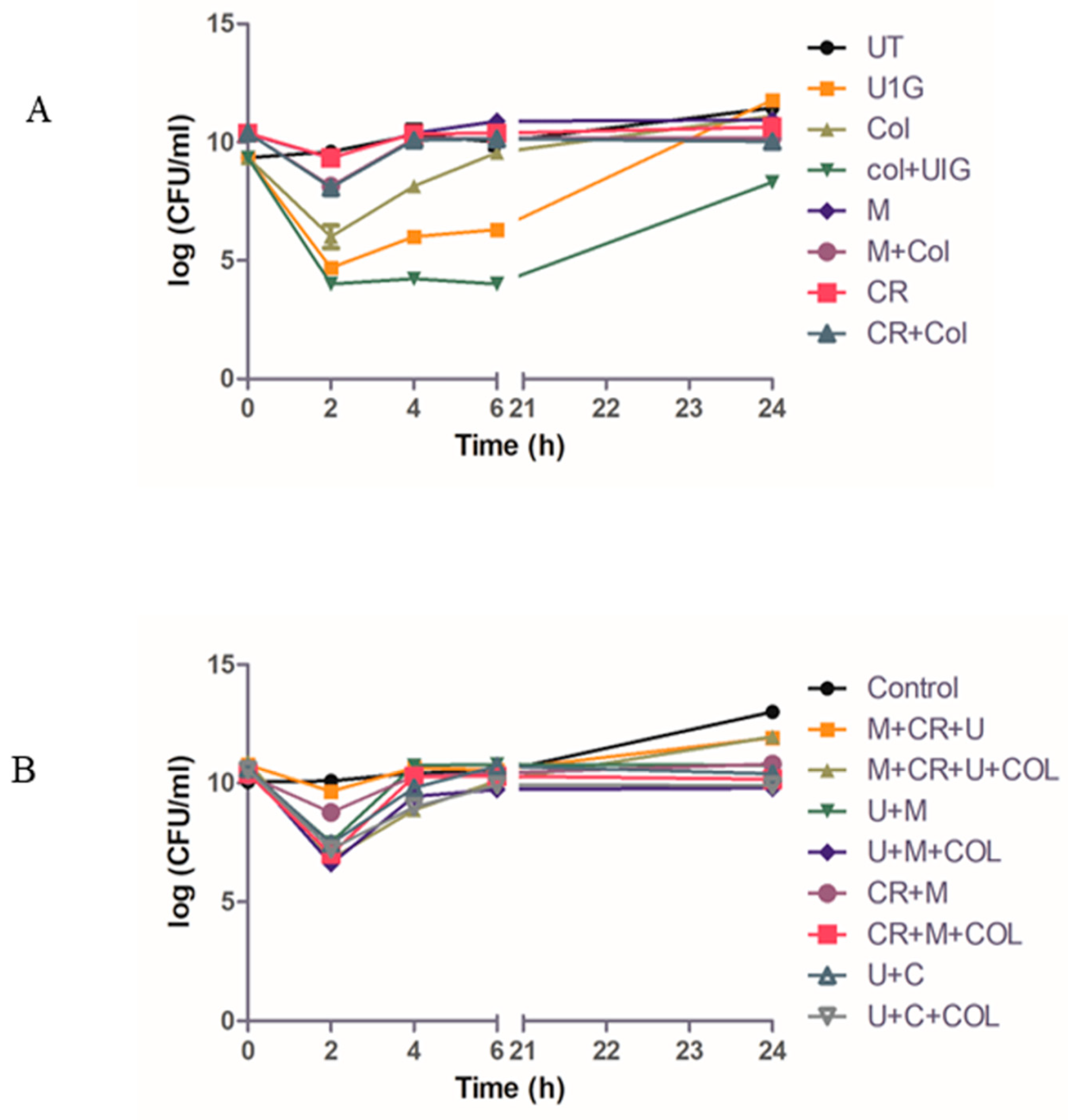
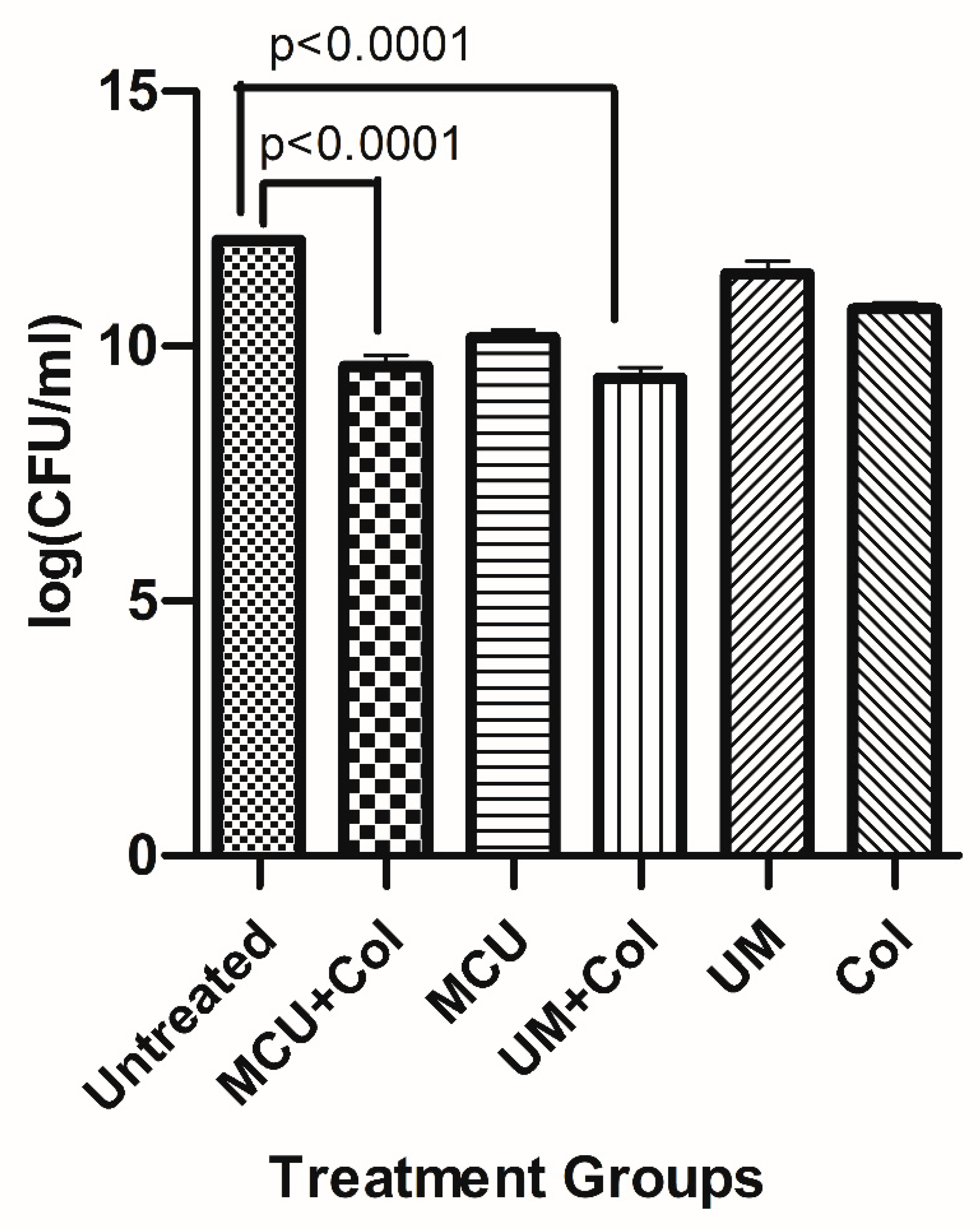
3.8. Time kill study:
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romero-Calle, D.; Benevides, R.G.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8. [Google Scholar] [CrossRef]
- Yin, Y.; Qiu, L.; Wang, G.; Guo, Z.; Wang, Z.; Qiu, J.; Li, R. Emergence and Transmission of Plasmid-Mediated Mobile Colistin Resistance Gene Mcr-10 in Humans and Companion Animals. Microbiol. Spectr. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Majewski, P.; Gutowska, A.; Smith, D.G.E.; Hauschild, T.; Majewska, P.; Hryszko, T.; Gizycka, D.; Kedra, B.; Kochanowicz, J.; Glowiński, J.; et al. Plasmid Mediated Mcr-1.1 Colistin-Resistance in Clinical Extraintestinal Escherichia Coli Strains Isolated in Poland. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Voelker, R. FDA Approves Bacteriophage Trial. JAMA 2019, 321, 638. [Google Scholar] [CrossRef] [PubMed]
- NIH No Title. 2020.
- NIH- Clinicaltrials No Title. 2020.
- Parfitt, T. Georgia: An Unlikely Stronghold for Bacteriophage Therapy. Lancet 2005, 365, 2166–2167. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage Therapy as a Potential Solution in the Fight against AMR: Obstacles and Possible Futures. Palgrave Commun. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Zaldastanishvili, E.; Leshkasheli, L.; Dadiani, M.; Nadareishvili, L.; Askilashvili, L.; Kvatadze, N.; Goderdzishvili, M.; Kutateladze, M.; Balarjishvili, N. Phage Therapy Experience at the Eliava Phage Therapy Center: Three Cases of Bacterial Persistence. Viruses 2021, 13, 1901. [Google Scholar] [CrossRef]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Wołkowicz, T.; et al. Characterization of a Bacteriophage, VB_Eco4M-7, That Effectively Infects Many Escherichia Coli O157 Strains. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Cepko, L.C.S.; Garling, E.E.; Dinsdale, M.J.; Scott, W.P.; Bandy, L.; Nice, T.; Faber-Hammond, J.; Mellies, J.L. Myoviridae Phage PDX Kills Enteroaggregative Escherichia Coli without Human Microbiome Dysbiosis. J. Med. Microbiol. 2020, 69, 309–323. [Google Scholar] [CrossRef]
- Wang, S.; Yin, B.; Yu, L.; Dang, M.; Guo, Z.; Yan, G.; Hu, D.; Gu, J.; Du, C.; Feng, X.; et al. Overexpression of AmpC Promotes Bacteriophage Lysis of Ampicillin-Resistant Escherichia Coli. Front. Microbiol. 2020, 10, 2973. [Google Scholar] [CrossRef]
- Valério, N.; Oliveira, C.; Jesus, V.; Branco, T.; Pereira, C.; Moreirinha, C.; Almeida, A. Effects of Single and Combined Use of Bacteriophages and Antibiotics to Inactivate Escherichia Coli. Virus Res. 2017, 240, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Champagne, C.P.; Gardner, N. The Spot Test Method for the In-Plant Enumeration of Bacteriophages with Paired Cultures of Lactobacillus Delbrueckii Subsp. Bulgaricus and Streptococcus Salivarius Subsp. Thermophilus. Int. Dairy J. 1995, 5, 417–425. [Google Scholar] [CrossRef]
- Adams, M. Assay of Phages by the Agar Layer Method. In Bacteriophages; Interscience Publishers, 1959; pp. 450–451.
- Sulcius, S.; Staniulis, J.; Paškauskas, R. Morphology and Distribution of Phage-like Particles in a Eutrophic Boreal Lagoon. Oceanologia 2011, 53, 587–603. [Google Scholar] [CrossRef]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage PhiYeO3-12, Specific for Yersinia Enterocolitica Serotype O:3, Is Related to Coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, Z.; Tian, C.; Chen, X.; Hu, L.; Wei, X.; Li, H.; Lin, W.; Jiang, A.; Feng, R.; et al. Characterizing the Biology of Lytic Bacteriophage VB_EaeM_φEap-3 Infecting Multidrug-Resistant Enterobacter Aerogenes. Front. Microbiol. 2019, 10, 420. [Google Scholar] [CrossRef]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage Therapy for Treatment of Virulent Klebsiella Pneumoniae Infection in a Mouse Model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Grillon, A.; Schramm, F.; Kleinberg, M.; Jehl, F. Comparative Activity of Ciprofloxacin, Levofloxacin and Moxifloxacin against Klebsiella Pneumoniae, Pseudomonas Aeruginosa and Stenotrophomonas Maltophilia Assessed by Minimum Inhibitory Concentrations and Time-Kill Studies. PLoS One 2016, 11, 1–10. [Google Scholar] [CrossRef]
- Audira, G.; Siregar, P.; Strungaru, S.-A.; Huang, J.-C.; Hsiao, C.-D. Which Zebrafish Strains Are More Suitable to Perform Behavioral Studies? A Comprehensive Comparison by Phenomic Approach. Biology (Basel). 2020, 9, 200. [Google Scholar] [CrossRef]
- Ganesan, A.; Christena, L.R.; Subbarao, H.M.V.; Venkatasubramanian, U.; Thiagarajan, R.; Sivaramakrishnan, V.; Kasilingam, K.; Saisubramanian, N.; Ganesan, S.S. Identification of Benzochromene Derivatives as a Highly Specific NorA Efflux Pump Inhibitor to Mitigate the Drug Resistant Strains of s. Aureus†. RSC Adv. 2016, 6, 30258–30267. [Google Scholar] [CrossRef]
- Neely, M.; Pfeifer, J.; Caparon, M. Streptococcus-Zebrafish Model of Bacterial Pathogenesis. Infect. Immun. 2002, 70, 3904–3914. [Google Scholar] [CrossRef] [PubMed]
- Boulé, J.; Sholberg, P.L.; Lehman, S.M.; O’Gorman, D.T.; Svircev, A.M. Isolation and Characterization of Eight Bacteriophages Infecting Erwinia Amylovora and Their Potential as Biological Control Agents in British Columbia, Canada. Can. J. Plant Pathol. 2011, 33, 308–317. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- M, J.; I, Z.; Y, R.; Y, M.; S, M.; TL, M. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36. [Google Scholar] [CrossRef]
- PP, C.; TM, L. TRNAscan-SE: Searching for TRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef]
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for Finding Prophage in Bacterial Genomes. Brief. Bioinform. 2017. [CrossRef]
- LM, R.-R.; KT, K. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes. 2016. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M. V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia Coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Boeckaerts, D.; Stock, M.; Criel, B.; Gerstmans, H.; De Baets, B.; Briers, Y. Predicting Bacteriophage Hosts Based on Sequences of Annotated Receptor-Binding Proteins. Sci. Reports 2021 111 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host Receptors for Bacteriophage Adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Chatterjee, S.; Rothenberg, E. Interaction of Bacteriophage λ with Its E. Coli Receptor, LamB. Viruses 2012, 4, 3162. [Google Scholar] [CrossRef]
- Sandulache, R.; Prehm, P.; Kamp, D. Cell Wall Receptor for Bacteriophage Mu G(+). J. Bacteriol. 1984, 160, 299. [Google Scholar] [CrossRef] [PubMed]
- Blagus, R.; Lusa, L. SMOTE for High-Dimensional Class-Imbalanced Data. BMC Bioinformatics 2013, 14, 1–16. [Google Scholar] [CrossRef]
- Pedregosa FABIANPEDREGOSA, F.; Michel, V.; Grisel OLIVIERGRISEL, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; Cournapeau, D.; Pedregosa, F.; Varoquaux, G.; et al. Scikit-Learn: Machine Learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Sundaramoorthy, N.S.; Mohan, H.M.; Subramaniam, S.; Raman, T.; Selva Ganesan, S.; Sivasubamanian, A.; Nagarajan, S. Ursolic Acid Inhibits Colistin Efflux and Curtails Colistin Resistant Enterobacteriaceae. AMB Express 2019, 9, 27. [Google Scholar] [CrossRef]
- J, W.; D, T.; AD, M.; P, M.; AM, K.; EM, A. From Orphan Phage to a Proposed New Family-the Diversity of N4-Like Viruses. Antibiot. (Basel, Switzerland) 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Puente, J.L.; Verdugo-Rodríguez, A.; Calva, E. Expression of Salmonella Typhi and Escherichia Coli OmpC Is Influenced Differently by Medium Osmolarity; Dependence on Escherichia Coli OmpR. Mol. Microbiol. 1991, 5, 1205–1210. [Google Scholar] [CrossRef]
- Sato, M.; Machida, K.; Arikado, E.; Saito, H.; Kakegawa, T.; Kobayashi, H. Expression of Outer Membrane Proteins in Escherichia Coli Growing at Acid PH. Appl. Environ. Microbiol. 2000, 66, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Mead, A.; Toutain, P.-L.; Richez, P.; Pelligand, L. Quantitative Pharmacodynamic Characterization of Resistance versus Heteroresistance of Colistin in E. Coli Using a Semimechanistic Modeling of Killing Curves. Antimicrob. Agents Chemother. 2022, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, X.; Zhao, L.; He, Y.; Yu, W.; Fu, S.; Ni, W.; Gao, Z. Heteroresistance Is Associated With in Vitro Regrowth During Colistin Treatment in Carbapenem-Resistant Klebsiella Pneumoniae. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage Therapy: An Alternative to Antibiotics in the Age of Multi-Drug Resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef]
- The Epic of Phage Therapy. In Proceedings of the Canadian Journal of Infectious Diseases and Medical Microbiology; Hindawi Limited, 2007; Volume 18, pp. 15–18.
- Chanishvili, N. Phage Therapy-History from Twort and d’Herelle Through Soviet Experience to Current Approaches. In Advances in Virus Research; Academic Press Inc., 2012; Volume 83, pp. 3–40.
- WHO No Title.
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages Mediate Defense against Phage Infection through Diverse Mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef]
- Sundaramoorthy, N.S.; Thothathri, S.; Bhaskaran, M.; GaneshPrasad, A.K.; Nagarajan, S. Phages from Ganges River Curtail in Vitro Biofilms and Planktonic Growth of Drug Resistant Klebsiella Pneumoniae in a Zebrafish Infection Model. AMB Express 2021, 11. [Google Scholar] [CrossRef]
- Dimri, A.; Awasthi, C.; Uniyal, S.; Nautiyal, A.; Singh, K.P. Isolation and Characterization of Coliform Bacteria and Bacteriophages from Ganga River in Northern Himalayan Regions. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1582–1592. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Nachimuthu, R. Therapeutic Characterization and Efficacy of Bacteriophage Cocktails Infecting Escherichia Coli, Klebsiella Pneumoniae, and Enterobacter Species. Front. Microbiol. 2019, 10, 574. [Google Scholar] [CrossRef]
- Khairnar, K. Ganges: Special at Its Origin. J. Biol. Res. 2016, 23, 16. [Google Scholar] [CrossRef]
- Kaur, R.; Yadav, B.; Tyagi, R.D. Microbiology of Hospital Wastewater. In Current Developments in Biotechnology and Bioengineering; Elsevier, 2020; pp. 103–148.
- Boyer, M.; Wisniewski-Dyé, F.; Combrisson, J.; Bally, R.; Duponnois, R.; Costechareyre, D. Nettle Manure: An Unsuspected Source of Bacteriophages Active against Various Phytopathogenic Bacteria. Arch. Virol. 2022, 167, 1099–1110. [Google Scholar] [CrossRef]
- Abedon, S.T.; Herschler, T.D.; Stopar, D. Bacteriophage Latent-Period Evolution as a Response to Resource Availability. Appl. Environ. Microbiol. 2001, 67, 4233–4241. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Hao, G.; Chen, A.I.; Liu, M.; Zhou, H.; Egan, M.; Yang, X.; Kan, B.; Wang, H.; Goulian, M.; Zhu, J. Colistin Resistance-Mediated Bacterial Surface Modification Sensitizes Phage Infection. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Versoza, C.J.; Pfeifer, S.P. Computational Prediction of Bacteriophage Host Ranges. Microorganisms 2022, 10, 149. [Google Scholar] [CrossRef]
- Qimron, U.; Marintcheva, B.; Tabor, S.; Richardson, C.C. Genomewide Screens for Escherichia Coli Genes Affecting Growth of T7 Bacteriophage. Proc. Natl. Acad. Sci. 2006, 103, 19039–19044. [Google Scholar] [CrossRef]
- Darcan, C.; Ozkanca, R.; Dil, Ö. The Role of RpoS, H-NS and AcP on the PH-Dependent OmpC and OmpF Porin Expressions of Escherichia Coli at Different PH. African J. Biotechnol. 2009, 8, 1845–1854. [Google Scholar]
- Heyde, M.; Portalier, R. Regulation of Major Outer Membrane Porin Proteins of Escherichia Coli K 12 by PH. Mol. Gen. Genet. MGG 1987, 208, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage Secondary Infection. Virol. Sin. 2015, 30, 3–10. [Google Scholar] [CrossRef]
- M, A.-Z.; M, W.; EK, C.; AF, G.; RE, S.; K, A.; A, A.; C, E.; C, M.; S, M.; et al. Identification of Novel Bacteriophages with Therapeutic Potential That Target Enterococcus Faecalis. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef]
- Cafora, M.; Deflorian, G.; Forti, F.; Ferrari, L.; Binelli, G.; Briani, F.; Ghisotti, D.; Pistocchi, A. Phage Therapy against Pseudomonas Aeruginosa Infections in a Cystic Fibrosis Zebrafish Model. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- M, E.; M, D.Z.; HJ, S. Application of Phage Therapy: Synergistic Effect of Phage EcSw (ΦEcSw) and Antibiotic Combination towards Antibiotic-Resistant Escherichia Coli. Transbound. Emerg. Dis. 2020, 67, 2809–2817. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.-F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): β-Lactam and Quinolone Antibiotics Stimulate Virulent Phage Growth. PLoS One 2007, 2, e799. [Google Scholar] [CrossRef] [PubMed]
- North, O.I.; Brown, E.D. Phage–Antibiotic Combinations: A Promising Approach to Constrain Resistance Evolution in Bacteria. Ann. N. Y. Acad. Sci. 2021, 1496, 23–34. [Google Scholar] [CrossRef]
- Maltas, J.; Wood, K.B. Pervasive and Diverse Collateral Sensitivity Profiles Inform Optimal Strategies to Limit Antibiotic Resistance. PLOS Biol. 2019, 17, e3000515. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
| All features | ANOVA Feature Selected Dataset | L2 Regularization Feature Selected Dataset | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Performance Score | RF | DT | LR | RF | DT | LR | RF | DT | LR |
| Precision | 0.93 | 0.88 | 0.91 | 0.86 | 0.85 | 0.85 | 0.84 | 0.64 | 0.78 |
| Recall | 0.90 | 0.86 | 0.91 | 0.86 | 0.83 | 0.83 | 0.79 | 0.66 | 0.77 |
| Accuracy | 0.90 | 0.84 | 0.90 | 0.86 | 0.83 | 0.83 | 0.79 | 0.66 | 0.77 |
| F-1 Score | 0.89 | 0.85 | 0.89 | 0.85 | 0.83 | 0.83 | 0.78 | 0.63 | 0.75 |
| MCC | 0.89 | 0.83 | 0.89 | 0.85 | 0.82 | 0.82 | 0.78 | 0.64 | 0.76 |
| Phage | RBP | Predicted Receptors | ||
|---|---|---|---|---|
| All Features | ANOVA | L2 Regularization | ||
| U1G phage | Phage tail spike protein (107 aa) |
LPS O antigen | LPS O antigen | OmpC or LamB |
| CR phage | Phage protein (828 aa) | FhuA | FhuA | OmpC |
| M phage | Phage protein (852 aa) | OmpC or LamB | FhuA and TonB | OmpF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





