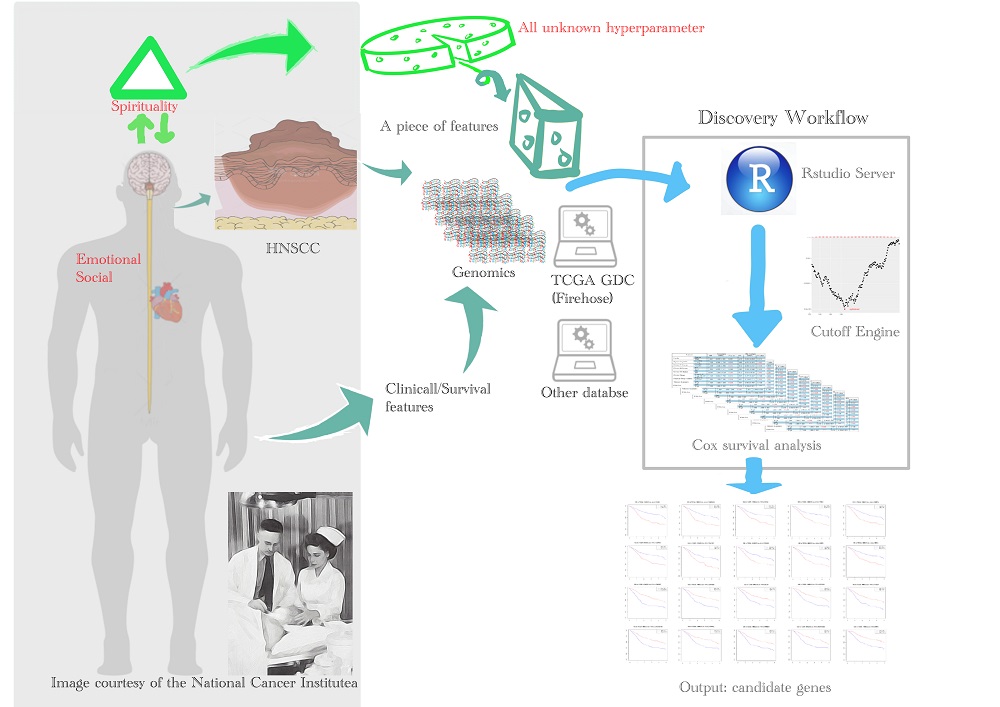
The survival analysis of the Cancer Genome Atlas (TCGA) dataset is a well-known method to discover the gene expression-based prognostic biomarkers of head and neck squamous cell carcinoma (HNSCC). A cutoff point is usually used in survival analysis for the patients' dichotomization in the continuous gene expression. There is some optimization software for cutoff determination. However, the software's predetermined cutoffs are usually set at the median or quantiles of gene expression value to perform the analyses. There are also few clinicopathological features available on their pre-processed data sets. We applied an in-house workflow, including data retrieving and pre-processing, feature selection, sliding-window cutoff selection, Kaplan-Meier survival analysis, and Cox proportional hazard modeling for biomarker discovery. In our approach for the TCGA HNSCC cohort, we scanned human protein-coding genes to find optimal cutoff values. After adjustment with confounders, the clinical tumor stage and the surgical margin involvement are independent risk factors for patients' prognosis. According to the resulting tables with Bonferroni-adjusted P value under the optimal cutoff and the hazard ratio, three biomarker candidates, CAMK2N1, CALML5, and FCGBP, are significantly associated with the patients' overall survival. We validated this discovery by using the other independent HNSCC dataset (GSE65858). Thus, we suggest the transcriptomic analysis could help for biomarker discovery.