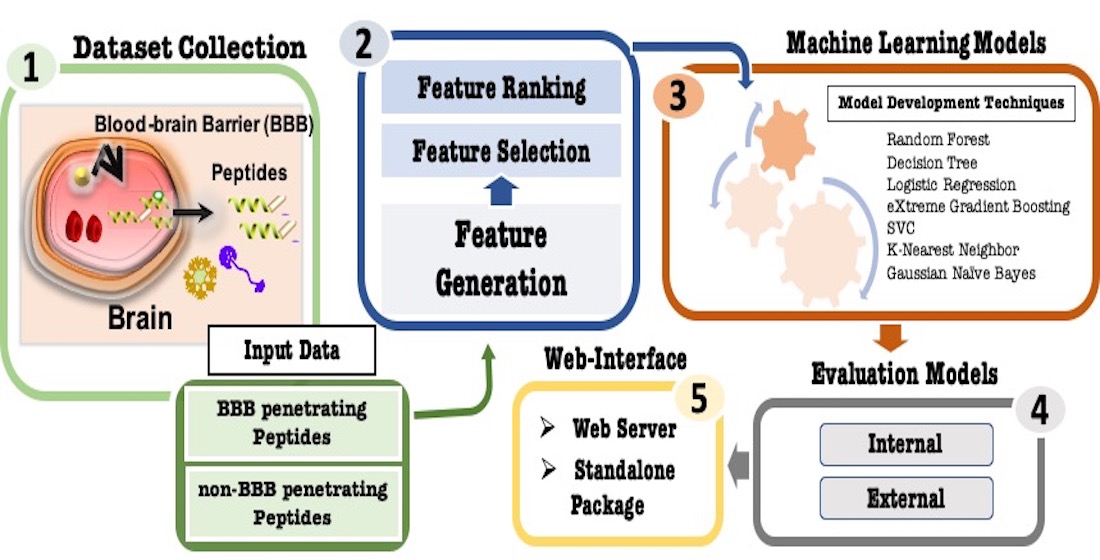
Blood-brain-barrier is a major obstacle in treating brain-related disorders as it does not allow to deliver drugs in the brain. In order to facilitate delivery of drugs in brain, we developed a method for predicting blood-brain-barrier penetrating peptides. These blood-brain barriers penetrating peptides (B3PPs) can act as therapeutic as well as drug delivery agents. We trained, tested, and evaluated our models on blood-brain-barrier peptides obtained from the B3Pdb database. First, we compute a wide range of peptide features then we select relevant peptide features. Finally, we developed numerous machine learning-based models for predicting blood-brain-barrier peptides using selected features. Our model based on random forest performed best on the top 80 selected features and achieved a maximum 85.08% accuracy with 0.93 AUROC. We also developed a web server, B3pred that implements our best models. It has three major modules that allow users to; i) predict B3PPs, ii) scanning B3PPs in a protein sequence, and iii) designing B3PPs using analogs. Our web server and standalone software is freely available at https://webs.iiitd.edu.in/raghava/b3pred/.