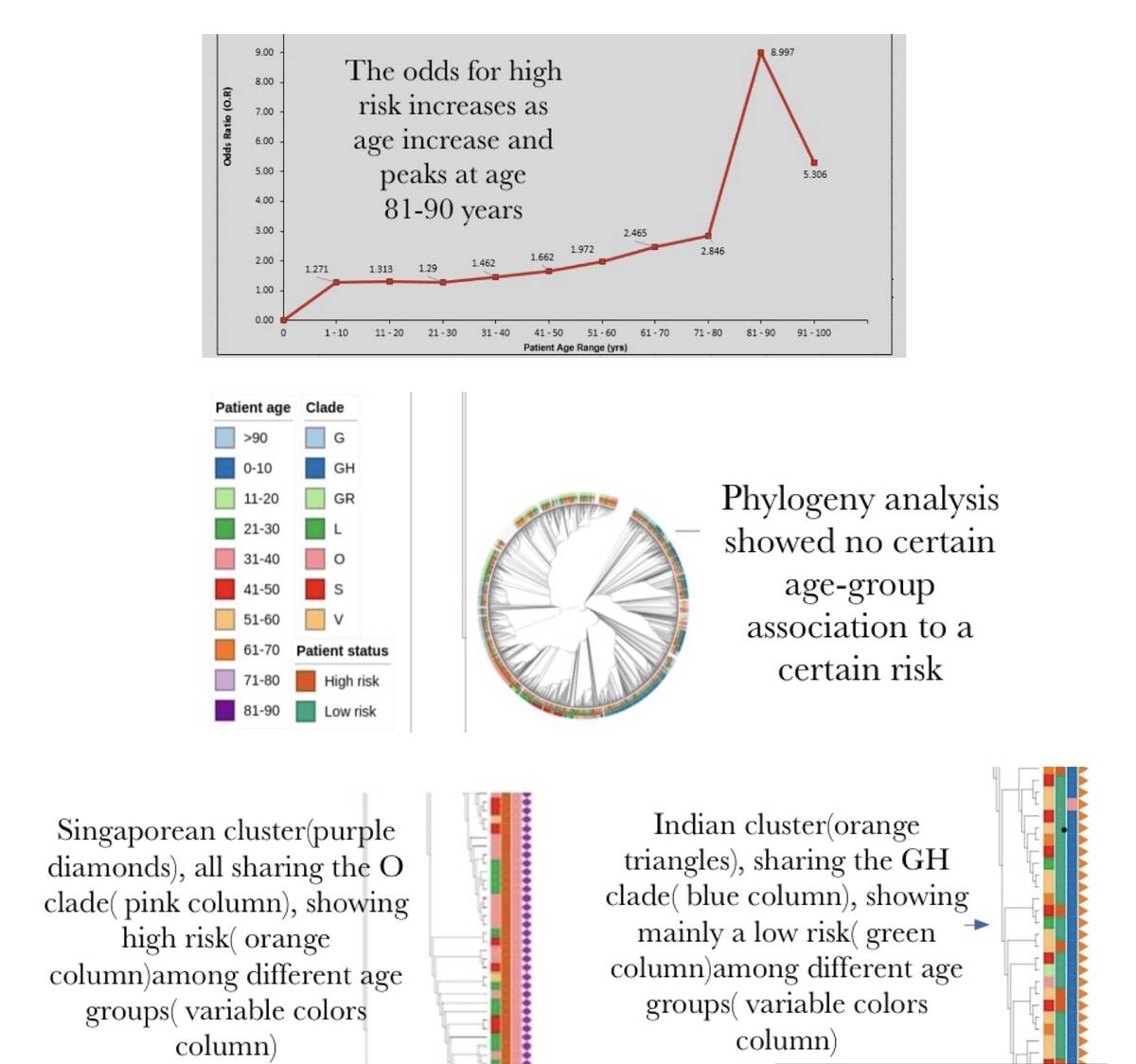
The age-related mortality and morbidity risk of COVID-19 has been considered speculative without enough scientific evidence. This study aimed to collect more evidence on the association between patient age and risk of severe disease state and/or mortality from SARS-CoV-2 infection. Genomic dataset along with metadata (3608 samples) retrieved from GISAID from different geographical regions were grouped into 10 age groups (0-10, 11-20, 21-30, 31-40, 41-50, 51-60, 61-70, 71-80, 81-90, 91-100 years) as well as high-risk or low-risk according to patient clinical status. Genomic sequences were aligned and analyzed using MAFFT and FASTTREE to build a phylogenetic tree in order to identify age-risk associations based on phylogenetic clustering. Case fatality rates (CFR), as well as the Odds ratio (OR) for high-risk outcomes, were calculated for different age groups. Results revealed that individuals aged between 25-50 years have the best immune response to the infection. On the other hand, disease fatality was higher in patients aging above 50 years. We created an application to calculate the OR of being at high risk given a certain age threshold from GISAID datasets. OR values increased between ages 1-10 years (1.271) and 11-20 years (1.313) but reduced at age range 21-30 years (1.290) and increased again for 61-70 years (2.465). CFR calculated for each of the age groups had peak values at 90-100 years (26.8%) and the lowest at 0-10 years (0%). The CFR for ages above 50 years was about twice greater (11.6%-26.8%) than that for ages below (0-6.6%). The phylogenetic analysis revealed that the majority of samples obtained from India showed low-risk among different age groups and were defined as clade GH. Another cluster from Singapore visualization showed unfavorable patient outcome across several age groups and were classified under clade O. To conclude, this study analyses showed a variety of age-risk associations. As scientists from different countries upload more genomes to globally shared databases, more evidence will reinforce mortality risk associations in COVID-19 patients.