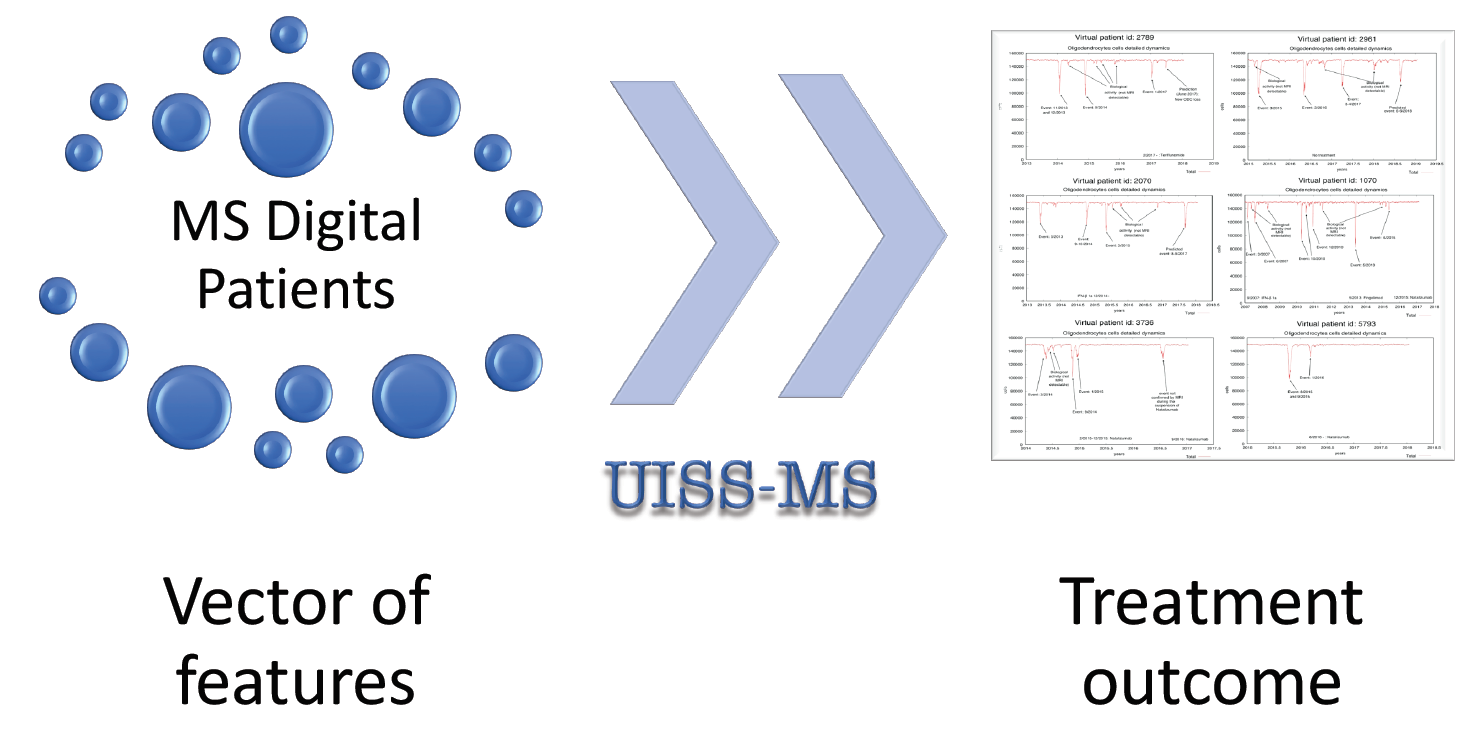
As of today, 20 disease modifying drugs (DMD) have been approved for the treatment of relapsing multiple sclerosis (MS) and, based on their efficacy, they can be grouped into moderate-efficacy DMDs and high-efficacy DMDs. The choice of the drug mostly relies on the judgement and experience of neurologists and the evaluation of therapeutic response can only be obtained by monitoring clinical and magnetic resonance imaging (MRI) status during follow up. In an era where therapies are focused on personalization, the aim of this study is to develop a modeling infrastructure to predict the evolution of relapsing MS and the response to treatments. We built a computational modeling infrastructure named UISS (Universal Immune System Simulator) able to simulate the main features and dynamics of the immune system activities. We extended UISS to simulate all the underlying MS pathogenesis and its interaction with the host immune system. This simulator is a multi-scale, multi-organ, agent based simulator with an attached module capable of simulating the dynamics of specific biological pathways at the molecular level. We simulated six MS patients with different relapsing-remitting courses. These patients were characterized on the basis of their age, sex, presence of oligoclonal bands, therapy and MRI lesion load at onset. The simulator framework is made freely available and can be used following the links provided in the availability section. Even though the model can be further personalized employing immunological parameters and genetic information, based on the available data we generated a few simulation scenarios for each patient, including those who matched the real clinical and MRI history. Moreover, for two patients, the simulator anticipated the timing of subsequent relapses, which really occurred, suggesting that UISS may have the potential to assist MS specialists in predicting the course of the disease and the response to treatment.