Submitted:
13 April 2025
Posted:
14 April 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Sample Collection and Strain Identification
Antimicrobial Susceptibility Testing
Plasmid Conjugation Assay
Whole genome Sequencing of blaNDM-Positive Strains
Bioinformatics Analysis of Assembled Genomes
Genetic Environment Analysis of blaNDM-Positive Strains
Statistical Analysis
Results
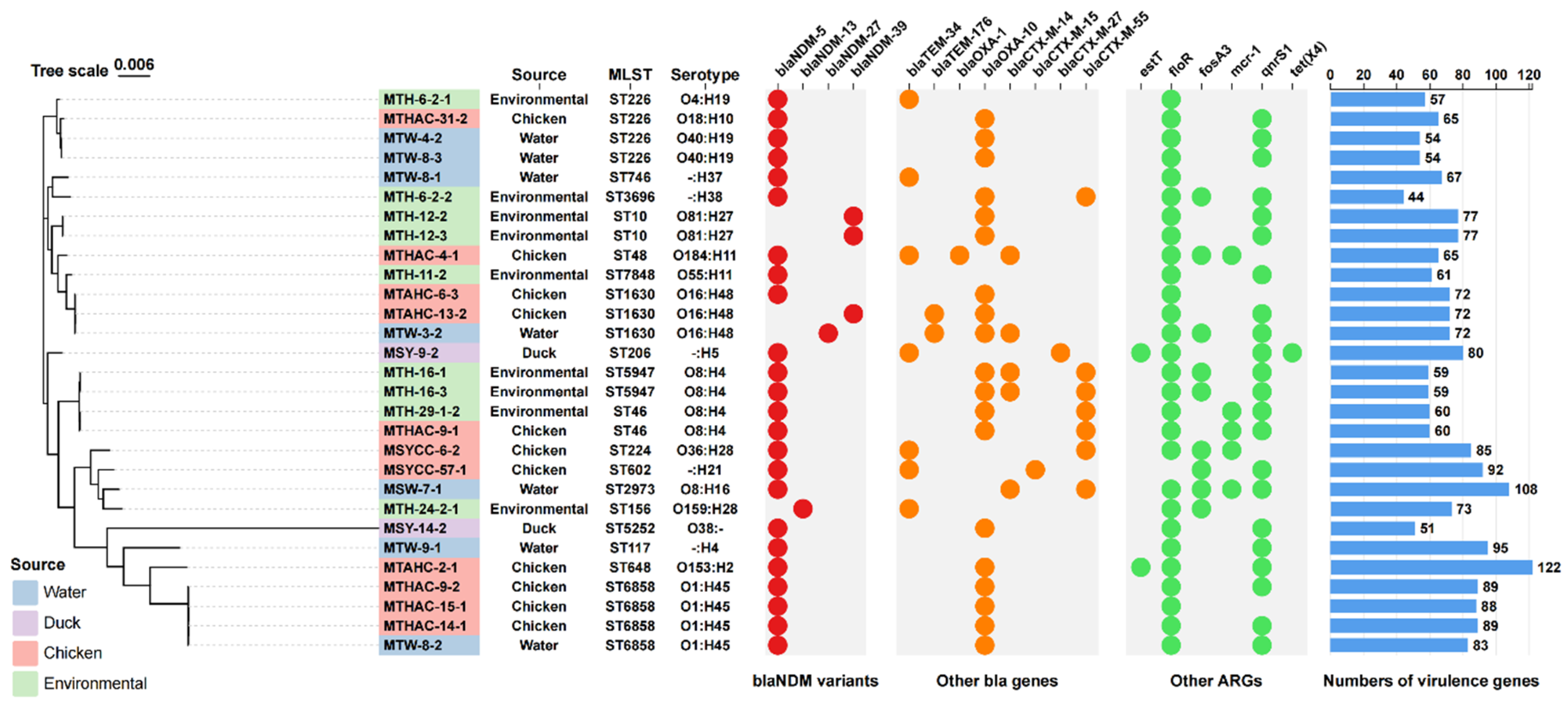
blaNDM-Positive Strains Profile
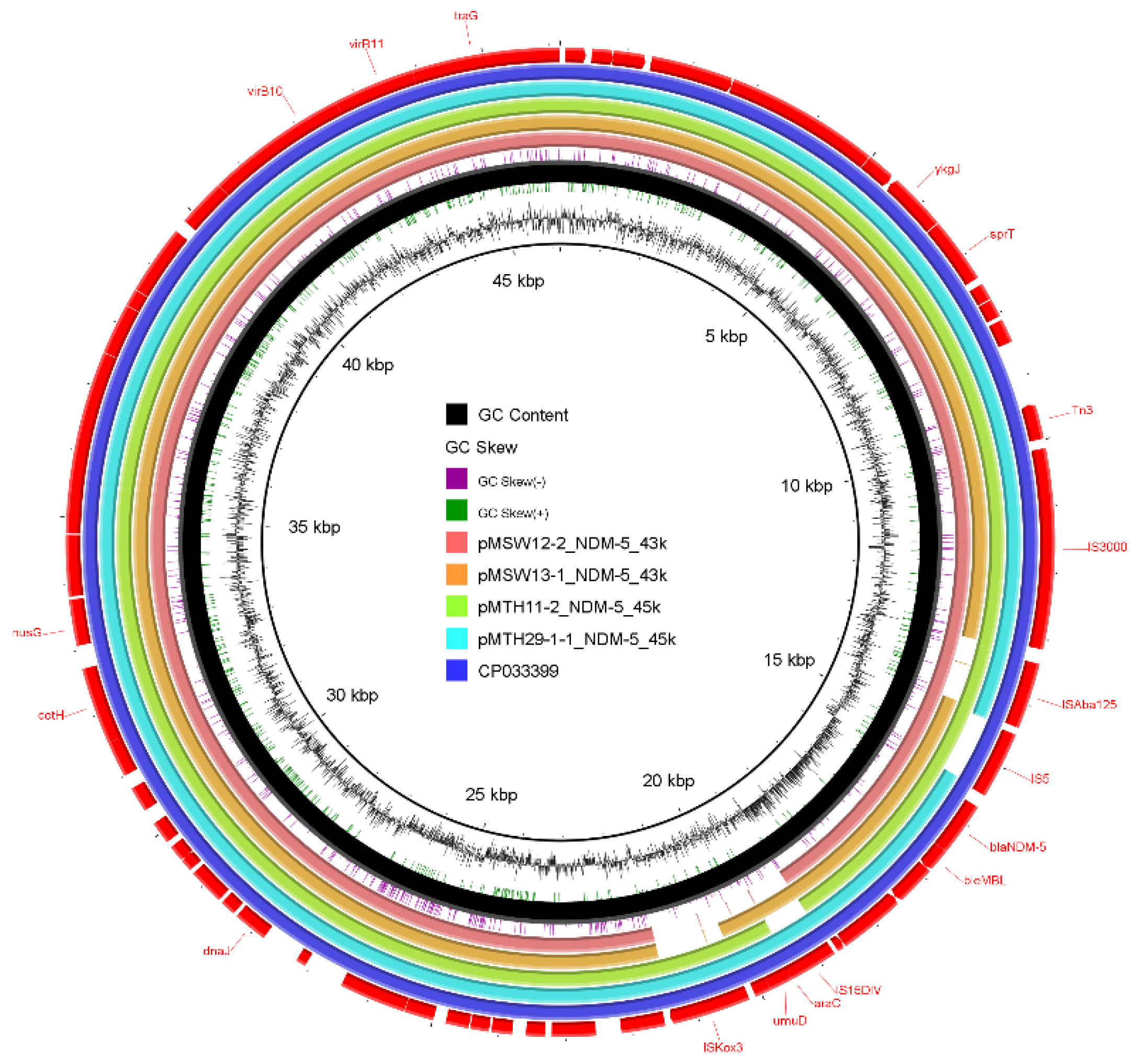
Genomic Analysis of blaNDM-Positive Strains
Genetic Environment Analysis of Various blaNDM Gene Variants
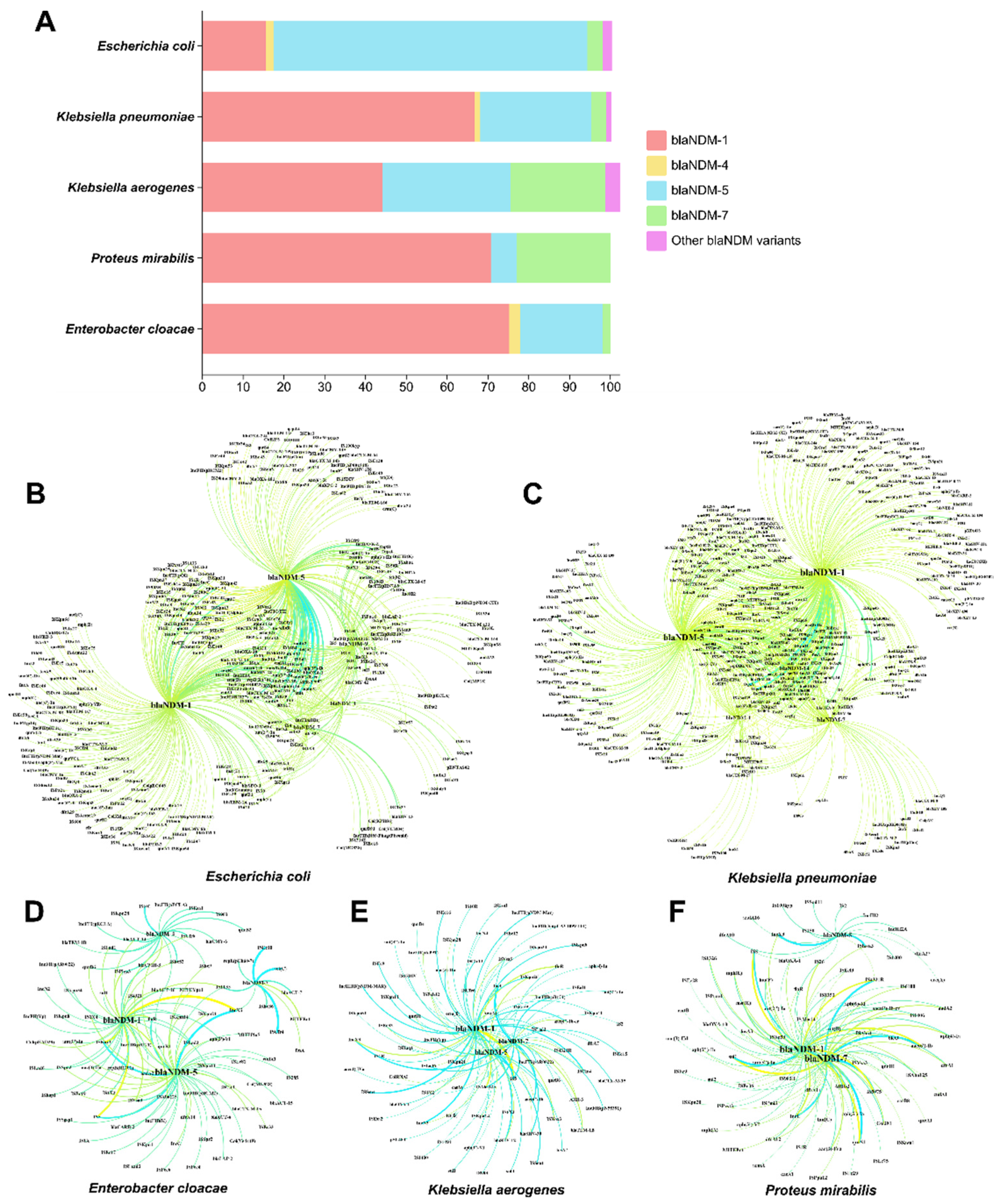
Correlation Analysis of blaNDM with Other ARGs, ISs and Plasmid Replicons
Discussion
Funding
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- ALAM M U, RAHMAN M, ABDULLAH AL M, et al. Human exposure to antimicrobial resistance from poultry production: Assessing hygiene and waste-disposal practices in Bangladesh [J]. International journal of hygiene and environmental health, 2019, 222(8): 1068-76. [CrossRef]
- WANG Y, HU Y, CAO J, et al. Antibiotic resistance gene reservoir in live poultry markets [J]. J Infect, 2019, 78(6): 445-53. [CrossRef]
- WANG Y, LYU N, LIU F, et al. More diversified antibiotic resistance genes in chickens and workers of the live poultry markets [J]. Environ Int, 2021, 153: 106534. [CrossRef]
- GAO X L, SHAO M F, LUO Y, et al. Airborne bacterial contaminations in typical Chinese wet market with live poultry trade [J]. Sci Total Environ, 2016, 572: 681-7. [CrossRef]
- TOO R J, KARIUKI S M, GITAO G C, et al. Carbapenemase-producing bacteria recovered from Nairobi River, Kenya surface water and from nearby anthropogenic and zoonotic sources [J]. PLoS One, 2024, 19(11): e0310026. [CrossRef]
- SU Y, XIN L, ZHANG F, et al. Drug resistance analysis of three types of avian-origin carbapenem-resistant Enterobacteriaceae in Shandong Province, China [J]. Poult Sci, 2023, 102(3): 102483. [CrossRef]
- YANG H, XIONG Z, CAO K, et al. Risk factors and molecular epidemiology of colonizing carbapenem-resistant Enterobacterales in pediatric inpatient in Shenzhen, China [J]. Journal of infection and public health, 2025, 18(1): 102614. [CrossRef]
- AL-MUSTAPHA A I, TIWARI A, LAUKKANEN-NINIOS R, et al. Wastewater based genomic surveillance key to population level monitoring of AmpC/ESBL producing Escherichia coli [J]. Sci Rep, 2025, 15(1): 7400. [CrossRef]
- LI C A, GUO C H, YANG T Y, et al. Whole-Genome Analysis of bla(NDM)-Bearing Proteus mirabilis Isolates and mcr-1-Positive Escherichia coli Isolates Carrying bla(NDM) from the Same Fresh Vegetables in China [J]. Foods (Basel, Switzerland), 2023, 12(3). [CrossRef]
- HUMPHRIES R, BOBENCHIK A M, HINDLER J A, et al. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition [J]. J Clin Microbiol, 2021, 59(12): e0021321. [CrossRef]
- CHEN Y, CHEN Y, SHI C, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data [J]. Gigascience, 2018, 7(1): 1-6. [CrossRef]
- BANKEVICH A, NURK S, ANTIPOV D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing [J]. Journal of computational biology : a journal of computational molecular cell biology, 2012, 19(5): 455-77. [CrossRef]
- ZANKARI E, HASMAN H, COSENTINO S, et al. Identification of acquired antimicrobial resistance genes [J]. J Antimicrob Chemother, 2012, 67(11): 2640-4. [CrossRef]
- SIGUIER P, PEROCHON J, LESTRADE L, et al. ISfinder: the reference centre for bacterial insertion sequences [J]. Nucleic Acids Res, 2006, 34(Database issue): D32-6. [CrossRef]
- CARATTOLI A, ZANKARI E, GARCíA-FERNáNDEZ A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing [J]. Antimicrob Agents Chemother, 2014, 58(7): 3895-903. [CrossRef]
- CHEN L, YANG J, YU J, et al. VFDB: a reference database for bacterial virulence factors [J]. Nucleic Acids Res, 2005, 33(Database issue): D325-8. [CrossRef]
- LIU M, LI X, XIE Y, et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements [J]. Nucleic Acids Res, 2019, 47(D1): D660-d5. [CrossRef]
- BESSONOV K, LAING C, ROBERTSON J, et al. ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data [J]. Microb Genom, 2021, 7(12). [CrossRef]
- SEEMANN T. Prokka: rapid prokaryotic genome annotation [J]. Bioinformatics (Oxford, England), 2014, 30(14): 2068-9. [CrossRef]
- PAGE A J, CUMMINS C A, HUNT M, et al. Roary: rapid large-scale prokaryote pan genome analysis [J]. Bioinformatics (Oxford, England), 2015, 31(22): 3691-3. [CrossRef]
- PRICE M N, DEHAL P S, ARKIN A P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix [J]. Molecular biology and evolution, 2009, 26(7): 1641-50. [CrossRef]
- ALIKHAN N F, PETTY N K, BEN ZAKOUR N L, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons [J]. BMC genomics, 2011, 12: 402. [CrossRef]
- LI Y, SUN X, DONG N, et al. Global distribution and genomic characteristics of carbapenemase-producing Escherichia coli among humans, 2005-2023 [J]. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy, 2024, 72: 101031. [CrossRef]
- CHKLOVSKI A, PARKS D H, WOODCROFT B J, et al. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning [J]. Nature methods, 2023, 20(8): 1203-12. [CrossRef]
- BASTIAN M, HEYMANN S, JACOMY M. Gephi: An Open Source Software for Exploring and Manipulating Networks [M]. 2009.
- YONG D, TOLEMAN M A, GISKE C G, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India [J]. Antimicrob Agents Chemother, 2009, 53(12): 5046-54. [CrossRef]
- DIEBOLD P J, RHEE M W, SHI Q, et al. Clinically relevant antibiotic resistance genes are linked to a limited set of taxa within gut microbiome worldwide [J]. Nat Commun, 2023, 14(1): 7366. [CrossRef]
- YAO J, HU Y, WANG X, et al. Carbapenem-resistant Morganella morganii carrying bla(KPC-2) or bla(NDM-1) in the clinic: one-decade genomic epidemiology analysis [J]. Microbiol Spectr, 2025: e0247624. [CrossRef]
- KE Y, ZHU Z, LU W, et al. Emerging bla(NDM)-positive Salmonella enterica in Chinese pediatric infections [J]. Microbiol Spectr, 2024, 12(12): e0148524. [CrossRef]
- ZHAO Q, GUO L, YE K, et al. Epidemiology, Phylogeny and Genetic Characterization of Carbapenem-resistant Citrobacter spp. from 5 hospitals in China [J]. J Glob Antimicrob Resist, 2025. [CrossRef]
- MARANO R B M, OSTER Y, BENENSON S, et al. An Omics-Guided Investigation of a Hospital Outbreak Caused by blaNDM-1-Producing Pseudocitrobacter faecalis [J]. The Journal of infectious diseases, 2025. [CrossRef]
- LIAN S, LIU C, CAI M, et al. Risk factors and molecular characterization of carbapenem resistant Escherichia coli recovered from a tertiary hospital in Fujian, China from 2021 to 2023 [J]. BMC Microbiol, 2024, 24(1): 374. [CrossRef]
- MORRIS R, WANG S. Building a pathway to One Health surveillance and response in Asian countries [J]. Science in One Health, 2024, 3: 100067. [CrossRef]
- GUAN Y, WANG Z, SHANG Z, et al. Steady existence of Escherichia coli co-resistant to carbapenem and colistin in an animal breeding area even after the colistin forbidden [J]. J Environ Manage, 2024, 371: 123084. [CrossRef]
- SHAFIQ M, ZENG M, PERMANA B, et al. Coexistence of bla (NDM-5) and tet(X4) in international high-risk Escherichia coli clone ST648 of human origin in China [J]. Front Microbiol, 2022, 13: 1031688. [CrossRef]
- WANG Y, WU C, ZHANG Q, et al. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin [J]. PLoS One, 2012, 7(5): e37152. [CrossRef]
- MEDUGU N, TICKLER I A, DURU C, et al. Phenotypic and molecular characterization of beta-lactam resistant Multidrug-resistant Enterobacterales isolated from patients attending six hospitals in Northern Nigeria [J]. Sci Rep, 2023, 13(1): 10306. [CrossRef]
- ZHANG F, LI Z, LIU X, et al. Carbapenem-resistant Citrobacter freundii harboring bla(KPC-2) and bla(NDM-1): a study on their transferability and potential dissemination via generating a transferrable hybrid plasmid mediated by IS6100 [J]. Front Microbiol, 2023, 14: 1239538. [CrossRef]
- PAJAND O, RAHIMI H, BADMASTI F, et al. Various arrangements of mobile genetic elements among CC147 subpopulations of Klebsiella pneumoniae harboring bla(NDM-1): a comparative genomic analysis of carbapenem resistant strains [J]. Journal of biomedical science, 2023, 30(1): 73. [CrossRef]
- OYELADE A A, IKHIMIUKOR O O, NWADIKE B I, et al. Assessing the risk of exposure to antimicrobial resistance at public beaches: Genome-based insights into the resistomes, mobilomes and virulomes of beta-lactams resistant Enterobacteriaceae from recreational beaches in Lagos, Nigeria [J]. International journal of hygiene and environmental health, 2024, 258: 114347. [CrossRef]
- ZI P, FANG M, YANG H, et al. Characterization of an NDM-1-Producing Citrobacter koseri Isolate from China [J]. Infect Drug Resist, 2024, 17: 61-7. [CrossRef]
- QIAN J, WU Z, ZHU Y, et al. One Health: a holistic approach for food safety in livestock [J]. Sci One Health, 2022, 1: 100015. [CrossRef]

| Strains | Source | Species | Conjugation recipient | MIC | |||||||
| MEM | IMP | AMP | CAZ | KAN | GEN | CIP | CL | ||||
| MTHAC-1-1 | Chicken | Escherichia coli | - | >128 | 8 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MTHAC-1-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 1 |
| MTHAC-2-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 2 | 0.5 | 0.5 |
| MTHAC-2-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | 4 | 1 | ≤0.25 |
| MTHAC-3-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 0.5 | ≤0.25 |
| MTHAC-3-3 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 16 | ≤0.25 |
| MTHAC-4-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 8 |
| MTHAC-4-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MTHAC-5-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 | ≤0.25 |
| MTHAC-6-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 |
| MTHAC-6-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 1 | 1 |
| MTHAC-8-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 |
| MTHAC-8-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | 32 | 16 | 0.5 | 1 |
| MTHAC-9-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 1 | 8 |
| MTHAC-9-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 | 1 |
| MTHAC-10-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 1 |
| MTHAC-10-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 1 | 0.5 |
| MTHAC-10-3 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 1 | ≤0.25 |
| MTHAC-11-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 4 | 1 |
| MTHAC-11-2 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MTHAC-12-1 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | >128 | 8 | ≤0.25 |
| MTHAC-12-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 2 | 0.5 |
| MTHAC-13-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | 2 |
| MTHAC-13-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 1 | ≤0.25 |
| MTHAC-14-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 |
| MTHAC-15-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 |
| MTHAC-15-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 2 | 0.5 |
| MTHAC-16-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 1 |
| MTHAC-16-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | 0.5 |
| MTHAC-17-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 2 |
| MTHAC-17-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 0.5 |
| MTHAC-17-3 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 1 |
| MTHAC-18-1 | Chicken | Escherichia coli | - | 64 | 128 | >128 | >128 | >128 | 128 | 8 | ≤0.25 |
| MTHAC-18-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | ≤0.25 |
| MTHAC-24-1 | Chicken | Escherichia coli | - | 64 | 128 | >128 | >128 | >128 | 64 | 0.5 | ≤0.25 |
| MTHAC-25-4 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 32 | 4 |
| MTHAC-27-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 2 |
| MTHAC-27-2 | Chicken | Escherichia coli | - | >128 | 4 | >128 | >128 | >128 | 128 | 8 | ≤0.25 |
| MTHAC-28-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 2 |
| MTHAC-31-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 16 |
| MTHAC-31-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 16 |
| MTAHC-1-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 2 | 4 |
| MTAHC-1-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 8 | 128 | 1 |
| MTAHC-2-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 4 | 8 |
| MTAHC-3-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 4 | 1 |
| MTAHC-4-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | 2 |
| MTAHC-4-3 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 4 |
| MTAHC-4-4 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 2 |
| MTAHC-5-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 32 | 1 |
| MTAHC-5-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 2 |
| MTAHC-6-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 1 |
| MTAHC-6-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 2 | 64 | ≤0.25 |
| MTAHC-6-3 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 4 |
| MTAHC-8-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 4 |
| MTAHC-12-1 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | >128 | 64 | 2 |
| MTAHC-12-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 64 | 0.5 |
| MTAHC-13-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 0.5 |
| MTAHC-13-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 4 |
| MTAHC-13-3 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 4 |
| MTAHC-14-3-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | - |
| MTAHC-14-3-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | - |
| MTAHC-15-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | - |
| MTAHC-15-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 8 | - |
| MTAHC-16-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 32 | - |
| MTAHC-16-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | - |
| MTAHC-16-3 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | - |
| MTH-1-3-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MTH-3-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | ≤0.25 |
| MTH-4-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 64 | 64 | ≤0.25 |
| MTH-4-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 64 | 64 | ≤0.25 |
| MTH-6-1-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MTH-6-1-2 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MTH-6-2-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 32 | >128 |
| MTH-6-2-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 32 | 2 | >128 |
| MTH-6-3-1 | Environmental | Klebsiella aerogenes | - | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 | >128 |
| MTH-6-3-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MTH-7-2-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | ≤0.25 |
| MTH-7-2-2 | Environmental | Klebsiella aerogenes | - | >128 | >128 | >128 | >128 | >128 | >128 | 32 | 128 |
| MTH-10-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MTH-11-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | 8 | 4 | 4 | 4 |
| MTH-11-2 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | 8 | 1 | 8 | 8 |
| MTH-12-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | 8 | 2 | 1 | 0.5 |
| MTH-12-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | 16 | 2 | 1 | >128 |
| MTH-12-3 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | 8 | 2 | 1 | >128 |
| MTH-13-2 | Environmental | Klebsiella pneumoniae | - | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| MTH-16-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | >128 |
| MTH-16-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 16 | 64 |
| MTH-16-3 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 32 | >128 |
| MTH-19-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 64 |
| MTH-24-2-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| MTH-26-1 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 4 | 2 | ≤0.25 |
| MTH-29-1-1 | Environmental | Enterobacter cloacae | C600 | >128 | >128 | >128 | >128 | 4 | 1 | ≤0.25 | >128 |
| MTH-29-1-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 1 | >128 |
| MTH-30-1-2 | Environmental | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MTW-1-1 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 |
| MTW-1-2 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 8 | ≤0.25 |
| MTW-2-1 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MTW-3-1 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | 128 | ≤0.25 |
| MTW-3-2 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| MTW-4-1 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 4 | >128 |
| MTW-4-2 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 4 | >128 |
| MTW-7-1 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 |
| MTW-8-1 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 2 | >128 |
| MTW-8-2 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 | >128 |
| MTW-8-3 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 64 | 4 | >128 |
| MTW-9-1 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 0.5 | >128 |
| MTW-18-1 | Water | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 64 | ≤0.25 |
| MTW-18-2 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 64 | 64 | 4 |
| MTW-18-3 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MTAHC18-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 64 | 64 | 2 |
| MTAHC18-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | 64 | ≤0.25 |
| MTNJC-3-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MTNJC-3-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 0.5 | ≤0.25 |
| MTNTC-2-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 1 | ≤0.25 |
| MTNTC-5-1 | Chicken | Klebsiella pneumoniae | - | >128 | >128 | >128 | >128 | >128 | 32 | 1 | ≤0.25 |
| MTNTC-5-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 1 | ≤0.25 |
| MTNTC-6-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | 0.5 | ≤0.25 |
| MTTZC-2-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 64 | 64 | ≤0.25 |
| MTTZC-2-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MTTZC-3-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MTAHG-11-1 | Pigeon | Providencia rettgeri | - | 32 | >128 | >128 | >128 | 16 | 32 | 8 | >128 |
| MTAHY-2-1 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MTAHY-4-1 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | ≤0.25 |
| MTAHY-4-2 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MTAHY-12-1 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | 8 |
| MTAHY-12-2 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 4 | ≤0.25 | ≤0.25 |
| MTAHY-13-1 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 32 | 1 | >128 |
| MTAHY-13-2 | Duck | Escherichia coli | - | >128 | >128 | >128 | >128 | 16 | 4 | ≤0.25 | ≤0.25 |
| MTYZG-24-3 | Pigeon | Klebsiella pneumoniae | - | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| MTYZG-33-1 | Pigeon | Klebsiella pneumoniae | - | 32 | 128 | >128 | 16 | 1 | ≤0.25 | ≤0.25 | ≤0.25 |
| MSYCC-4-2 | Chicken | Escherichia coli | C600 | 2 | 128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSYCC-5-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | 8 | 1 | ≤0.25 | ≤0.25 |
| MSYCC-6-1 | Chicken | Escherichia coli | C600 | 128 | >128 | >128 | >128 | 16 | 4 | 32 | ≤0.25 |
| MSYCC-6-2 | Chicken | Escherichia coli | - | 32 | 128 | >128 | >128 | >128 | >128 | 8 | 4 |
| MSYCC-11-1 | Chicken | Escherichia coli | - | 8 | >128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 |
| MSYCC-11-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MSYCC-13-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSYCC-13-2 | Chicken | Escherichia coli | C600 | 2 | >128 | >128 | >128 | >128 | 32 | ≤0.25 | ≤0.25 |
| MSYCC-19-1 | Chicken | Escherichia coli | C600 | 128 | >128 | >128 | >128 | >128 | >128 | 1 | ≤0.25 |
| MSYCC-20-1 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSYCC-20-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MSYCC-21-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 0.5 | ≤0.25 |
| MSYCC-21-2 | Chicken | Escherichia coli | C600 | 32 | >128 | >128 | >128 | >128 | >128 | 0.5 | ≤0.25 |
| MSYCC-24-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MSYCC-25-1 | Chicken | Escherichia coli | C600 | 32 | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MSYCC-25-2 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 16 | 0.5 |
| MSYCC-28-2 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MSYCC-31-1 | Chicken | Escherichia coli | - | 32 | 128 | >128 | >128 | >128 | 128 | 32 | ≤0.25 |
| MSYCC-32-1 | Chicken | Escherichia coli | C600 | 16 | >128 | >128 | >128 | >128 | 32 | ≤0.25 | 1 |
| MSYCC-34-1 | Chicken | Escherichia coli | - | 16 | >128 | >128 | >128 | >128 | >128 | ≤0.25 | 2 |
| MSYCC-34-2 | Chicken | Escherichia coli | C600 | 4 | 32 | >128 | >128 | >128 | 64 | 8 | ≤0.25 |
| MSYCC-35-1 | Chicken | Escherichia coli | C600 | 32 | >128 | >128 | >128 | >128 | >128 | 0.5 | 0.5 |
| MSYCC-37-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | 64 | 32 | ≤0.25 |
| MSYCC-40-1 | Chicken | Escherichia coli | C600 | 128 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MSYCC-42-1 | Chicken | Escherichia coli | C600 | 16 | 16 | >128 | >128 | >128 | 1 | ≤0.25 | ≤0.25 |
| MSYCC-43-1 | Chicken | Escherichia coli | C600 | 128 | 128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MSYCC-45-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | 32 | 32 | ≤0.25 |
| MSYCC-45-2 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | 64 | ≤0.25 | ≤0.25 |
| MSYCC-46-1 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | >128 | 32 | ≤0.25 |
| MSYCC-47-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MSYCC-48-1 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | 2 | 2 | ≤0.25 |
| MSYCC-49-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | 16 | ≤0.25 |
| MSYCC-51-1 | Chicken | Escherichia coli | - | 128 | >128 | >128 | >128 | >128 | >128 | 2 | ≤0.25 |
| MSYCC-51-2 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 |
| MSYCC-51-3 | Chicken | Escherichia coli | C600 | 32 | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MSYCC-52-1 | Chicken | Escherichia coli | C600 | 64 | >128 | >128 | >128 | >128 | 128 | 32 | 0.5 |
| MSYCC-52-2 | Chicken | Escherichia coli | C600 | 128 | >128 | >128 | >128 | >128 | >128 | 16 | 1 |
| MSYCC-54-1 | Chicken | Escherichia coli | C600 | 32 | >128 | >128 | >128 | >128 | 2 | 0.5 | ≤0.25 |
| MSYCC-54-2 | Chicken | Proteus mirabilis | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 16 | >128 |
| MSYCC-55-1 | Chicken | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 64 | 1 |
| MSYCC-57-1 | Chicken | Escherichia coli | - | >128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 | 8 |
| MSNTC-3-1 | Chicken | Escherichia coli | C600 | 64 | 128 | >128 | >128 | 4 | 2 | 16 | ≤0.25 |
| MSNTC-4-1 | Chicken | Escherichia coli | C600 | 8 | 64 | >128 | >128 | >128 | 128 | 8 | ≤0.25 |
| MSNTC-7-1 | Chicken | Escherichia coli | C600 | 64 | 64 | >128 | >128 | 4 | 1 | >128 | ≤0.25 |
| MSNTC-9-1 | Chicken | Escherichia coli | C600 | 32 | 64 | >128 | >128 | >128 | 128 | >128 | ≤0.25 |
| MSNTC-10-1 | Chicken | Escherichia coli | - | 32 | 64 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MSNTC-10-2 | Chicken | Escherichia coli | C600 | 64 | 128 | >128 | >128 | >128 | >128 | ≤0.25 | ≤0.25 |
| MSNTC-12-1 | Chicken | Escherichia coli | C600 | 64 | 128 | >128 | >128 | >128 | 128 | 64 | ≤0.25 |
| MSNTC-12-2 | Chicken | Escherichia coli | - | 64 | >128 | >128 | >128 | >128 | >128 | >128 | ≤0.25 |
| MSNTC-13-1 | Chicken | Escherichia coli | C600 | 2 | >128 | >128 | >128 | 4 | 2 | 32 | ≤0.25 |
| MSNTC-13-2 | Chicken | Escherichia coli | C600 | 32 | 64 | >128 | >128 | 4 | 2 | 16 | ≤0.25 |
| MSNTC-14-1 | Chicken | Escherichia coli | C600 | 16 | 64 | >128 | >128 | >128 | 1 | 8 | ≤0.25 |
| MSNTC-14-2 | Chicken | Escherichia coli | C600 | 32 | 64 | >128 | >128 | >128 | 2 | 8 | ≤0.25 |
| MSNTC-17-1 | Chicken | Escherichia coli | C600 | 32 | 64 | >128 | >128 | >128 | 1 | ≤0.25 | ≤0.25 |
| MSNTC-21-1 | Chicken | Escherichia coli | C600 | 4 | 8 | >128 | >128 | >128 | >128 | 8 | ≤0.25 |
| MSNTC-21-2 | Chicken | Escherichia coli | C600 | 4 | 8 | >128 | >128 | >128 | >128 | 4 | ≤0.25 |
| MSNTC-29-1 | Chicken | Escherichia coli | C600 | 64 | 128 | >128 | >128 | 4 | 1 | 16 | ≤0.25 |
| MSY-2-1 | Duck | Escherichia coli | - | 64 | 64 | >128 | >128 | 8 | 32 | 64 | ≤0.25 |
| MSY-2-2 | Duck | Escherichia coli | - | 64 | 128 | >128 | >128 | >128 | 128 | 0.5 | ≤0.25 |
| MSY-3-1 | Duck | Escherichia coli | - | 64 | 64 | >128 | >128 | 16 | 32 | 32 | ≤0.25 |
| MSY-3-2 | Duck | Escherichia coli | C600 | 8 | 16 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSY-4-1 | Duck | Escherichia coli | - | 32 | 64 | >128 | >128 | 8 | 32 | 64 | ≤0.25 |
| MSY-4-2 | Duck | Escherichia coli | - | ≤0.25 | 8 | >128 | >128 | >128 | 16 | ≤0.25 | ≤0.25 |
| MSY-6-2 | Duck | Escherichia coli | - | 2 | 8 | >128 | >128 | >128 | 64 | ≤0.25 | ≤0.25 |
| MSY-7-1 | Duck | Escherichia coli | - | ≤0.25 | 1 | >128 | >128 | >128 | 8 | ≤0.25 | ≤0.25 |
| MSY-8-1 | Duck | Escherichia coli | C600 | 32 | 128 | >128 | >128 | >128 | 64 | ≤0.25 | ≤0.25 |
| MSY-9-2 | Duck | Escherichia coli | C600 | 64 | 64 | >128 | >128 | 128 | 64 | 1 | >128 |
| MSY-10-1 | Duck | Escherichia coli | - | 32 | 128 | >128 | >128 | 4 | 1 | 16 | ≤0.25 |
| MSY-10-2 | Duck | Escherichia coli | C600 | 16 | 32 | >128 | >128 | >128 | 64 | ≤0.25 | ≤0.25 |
| MSY-11-1 | Duck | Escherichia coli | C600 | 32 | 32 | >128 | >128 | >128 | 16 | 4 | ≤0.25 |
| MSY-13-1 | Duck | Escherichia coli | C600 | 128 | 128 | >128 | >128 | >128 | 64 | 8 | ≤0.25 |
| MSY-13-2 | Duck | Escherichia coli | C600 | 2 | 8 | >128 | >128 | >128 | 64 | ≤0.25 | ≤0.25 |
| MSY-14-1 | Duck | Escherichia coli | - | 32 | 64 | >128 | >128 | 16 | 32 | 64 | ≤0.25 |
| MSY-14-2 | Duck | Escherichia coli | - | 128 | 128 | >128 | >128 | >128 | >128 | 1 | >128 |
| MSY-15-1 | Duck | Escherichia coli | C600 | 128 | 128 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSG-2-1 | Pigeon | Escherichia coli | - | 64 | 64 | >128 | >128 | >128 | 1 | 8 | ≤0.25 |
| MSG-4-1 | Pigeon | Escherichia coli | - | 32 | 128 | >128 | >128 | >128 | >128 | 64 | ≤0.25 |
| MSG-5-1 | Pigeon | Escherichia coli | C600 | 128 | 128 | >128 | >128 | >128 | 64 | 64 | ≤0.25 |
| MSE-1-1 | Goose | Escherichia coli | - | 1 | 8 | >128 | >128 | >128 | 16 | ≤0.25 | ≤0.25 |
| MSE-2-1 | Goose | Escherichia coli | - | 64 | 128 | >128 | >128 | >128 | 16 | 2 | ≤0.25 |
| MSW-2-1 | Water | Escherichia coli | C600 | 16 | 8 | >128 | >128 | >128 | >128 | 8 | 2 |
| MSW-4-1 | Water | Klebsiella pneumoniae | - | 32 | 16 | >128 | >128 | >128 | 0.5 | 0.5 | ≤0.25 |
| MSW-4-2 | Water | Escherichia coli | - | 16 | 16 | >128 | >128 | 8 | 2 | 0.5 | ≤0.25 |
| MSW-5-1 | Water | Escherichia coli | C600 | 16 | 8 | >128 | >128 | 8 | 1 | ≤0.25 | ≤0.25 |
| MSW-5-2 | Water | Escherichia coli | - | 8 | 4 | >128 | >128 | >128 | 32 | 16 | ≤0.25 |
| MSW-6-1 | Water | Escherichia coli | C600 | 64 | 8 | >128 | >128 | >128 | 128 | ≤0.25 | ≤0.25 |
| MSW-7-1 | Water | Escherichia coli | C600 | 64 | 8 | >128 | >128 | >128 | 32 | 128 | 4 |
| MSW-7-2 | Water | Klebsiella pneumoniae | - | 32 | 16 | >128 | >128 | >128 | 64 | 0.5 | ≤0.25 |
| MSW-9-1 | Water | Escherichia coli | C600 | 32 | 8 | >128 | >128 | >128 | 16 | 1 | ≤0.25 |
| MSW-11-1 | Water | Escherichia coli | - | 32 | 16 | >128 | >128 | >128 | 2 | ≤0.25 | ≤0.25 |
| MSW-11-2 | Water | Escherichia coli | C600 | 8 | 16 | >128 | >128 | >128 | 128 | 0.5 | ≤0.25 |
| MSW-12-1 | Water | Escherichia coli | C600 | 32 | 16 | >128 | >128 | 8 | 2 | 8 | ≤0.25 |
| MSW-12-2 | Water | Enterobacter cloacae | C600 | 32 | 16 | >128 | >128 | 4 | 1 | ≤0.25 | ≤0.25 |
| MSW-13-1 | Water | Enterobacter cloacae | C600 | >128 | >128 | >128 | >128 | 1 | 0.5 | ≤0.25 | ≤0.25 |
| MSW-13-2 | Water | Escherichia coli | C600 | 64 | 16 | >128 | >128 | 4 | 0.5 | 16 | ≤0.25 |
| MSW-14-1 | Water | Escherichia coli | - | 32 | 8 | >128 | >128 | 8 | ≤0.25 | 8 | ≤0.25 |
| MSW-15-1 | Water | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | ≤0.25 |
| MSH-11-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | >128 | 128 | 1 | ≤0.25 |
| MSH-14-1 | Environmental | Enterobacter cloacae | - | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 |
| MSH-17-1 | Environmental | Escherichia coli | C600 | >128 | >128 | >128 | >128 | 64 | 128 | 64 | ≤0.25 |
| MSH-19-1 | Environmental | Klebsiella pneumoniae | - | >128 | >128 | >128 | >128 | >128 | >128 | 8 | ≤0.25 |
| MSH-19-2 | Environmental | Escherichia coli | - | 64 | 16 | >128 | >128 | >128 | 8 | 1 | ≤0.25 |
| MSH-21-1 | Environmental | Escherichia coli | C600 | 8 | 4 | >128 | >128 | >128 | 16 | 2 | ≤0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





