Submitted:
15 December 2024
Posted:
16 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
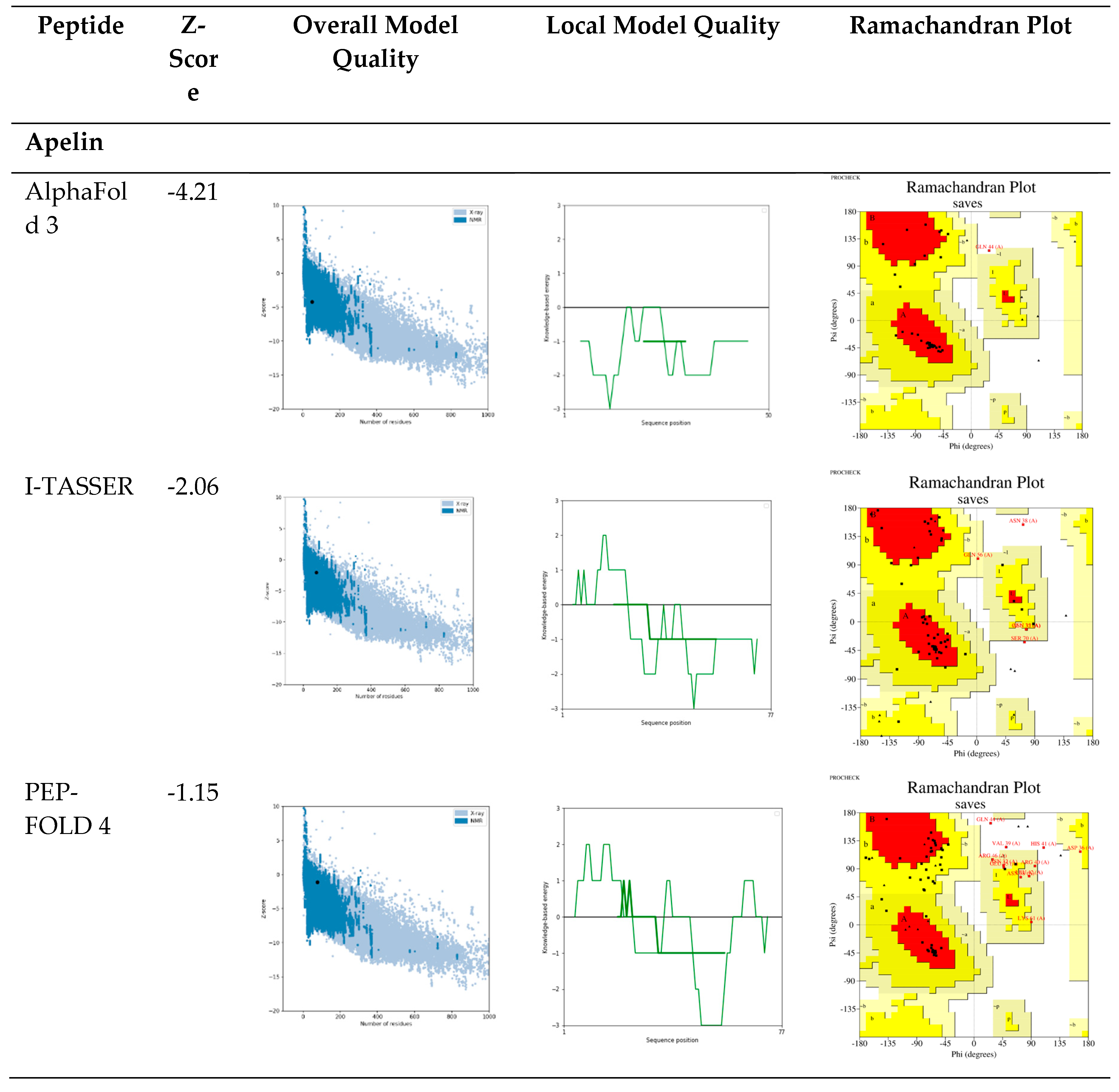
2.1. Evaluation of Structure Prediction Tools
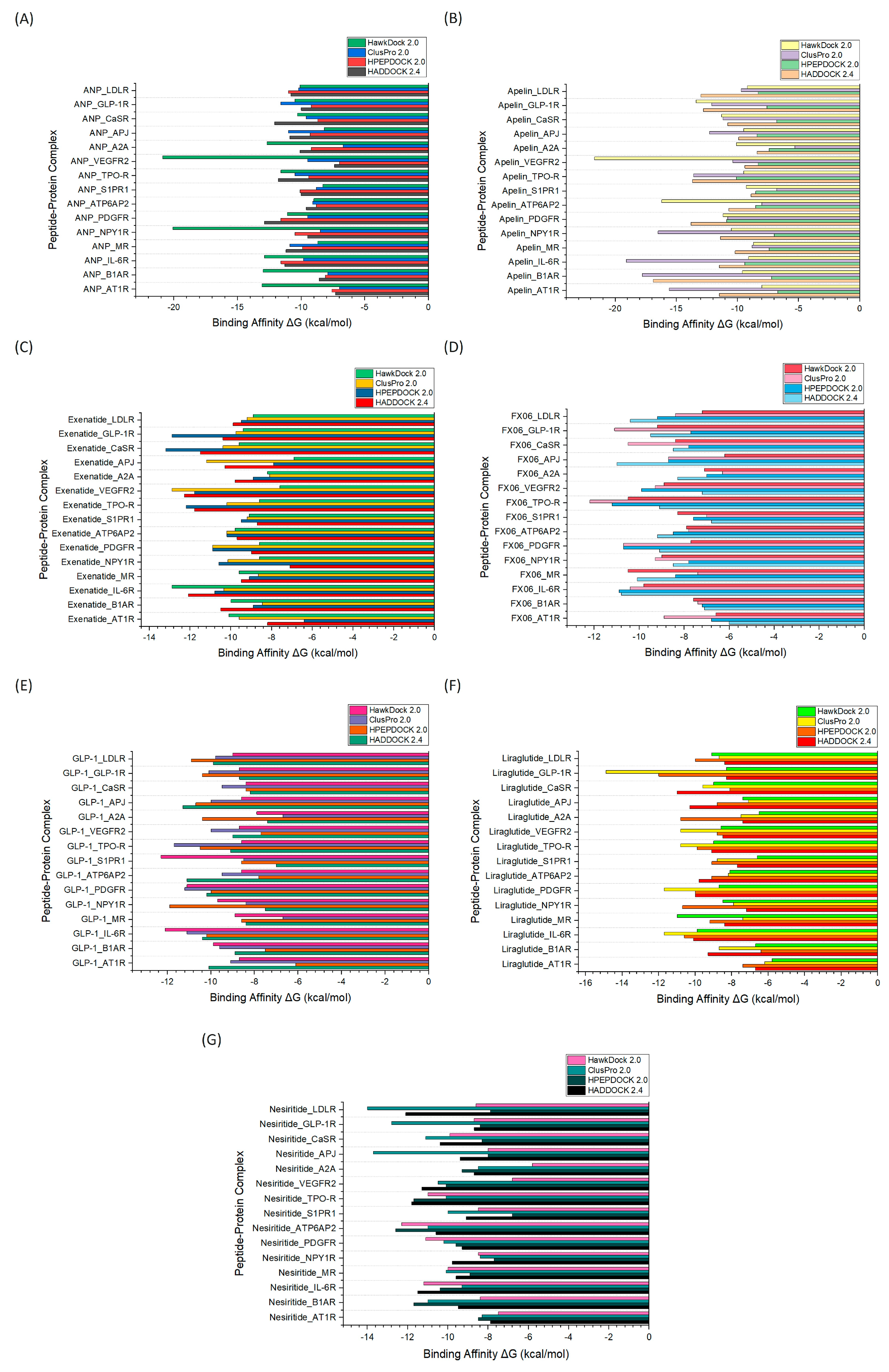
2.2. Evaluation of Molecular Docking Tools
2.3. Molecular Dynamics (MD) Simulation
| Therapeutic Peptide-Protein Complex | Average RMSD (Å) | Average RMSF (Å) | Average RoG (Å) | Number of hydrogen bonds between the two proteins |
| Apelin_AT1R | 2.124 | 1.821 | 2.352 | 8 |
| Apelin_ β1AR | 2.267 | 1.744 | 2.342 | 10 |
| Apelin_ IL-6R | 2.011 | 1.637 | 2.346 | 9 |
| Liraglutide_MR | 2.987 | 2.378 | 2.632 | 5 |
| ANP_ NPY1R | 2.824 | 2.412 | 2.639 | 6 |
| Apelin_ PDGFR | 2.357 | 2.004 | 2.499 | 7 |
| Apelin_ ATP6AP2 | 2.139 | 1.904 | 2.519 | 8 |
| ANP_ S1PR1 | 2.902 | 2.554 | 2.652 | 4 |
| FX06_ TPO-R | 2.706 | 2.228 | 2.629 | 5 |
| Apelin_VEGFR2 | 2.008 | 1.511 | 2.326 | 11 |
| ANP_ A2A | 2.675 | 2.425 | 2.653 | 6 |
| Apelin_APJ | 2.232 | 1.867 | 2.446 | 10 |
| Exenatide_CaSR | 3.012 | 2.708 | 2.691 | 4 |
| Apelin_GLP-1R | 2.356 | 1.912 | 2.487 | 9 |
| ANP_LDLR | 2.899 | 2.509 | 2.624 | 5 |
2.4. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
| Therapeutic Peptide-Protein Complex |
MM/PBSA Calculation Results ΔGbinding (kcal/mol) |
| Apelin_AT1R | -140.54 |
| Apelin_ β1AR | -156.53 |
| Apelin_ IL-6R | -163.66 |
| Liraglutide_MR | -50.27 |
| ANP_ NPY1R | -80.38 |
| Apelin_ PDGFR | -81.69 |
| Apelin_ ATP6AP2 | -103.43 |
| ANP_ S1PR1 | -54.57 |
| FX06_ TPO-R | -60.90 |
| Apelin_VEGFR2 | -199.17 |
| ANP_ A2A | -87.74 |
| Apelin_APJ | -171.62 |
| Exenatide_CaSR | -49.35 |
| Apelin_GLP-1R | -96.70 |
| ANP_LDLR | -49.26 |
3. Discussion
4. Limitations, Clinical Implications, and Future Works
4.1. Limitations
4.2. Clinical Implications
4.3. Future Works
5. Materials and Methods
5.1. Therapeutic Peptides
| Therapeutic Peptide | Sequence |
Binding Site (Position of Residues) |
| ANP | SLRRSSCFGGRMDRIGAQSGLGCNSFRY | 4, 5, 6, 7, 8, 10, 11, 12, 14, 15, 23, 24, 26 |
| Apelin | MNLRLCVQALLLLWLSLTAVCGGSLMPLPD | 10, 11, 14, 15, 25, 26, 27, 28, 29 |
| Exenatide | HGEGTFTSDLSKQMEEEAVRLFIEWLKNGG | 14, 17, 18, 21 |
| FX06 | MKHLLLLLLCVFLVKSQGVNDNEEGFFS | 13, 17, 19, 24 |
| GLP-1 | HAEGTFTTSDVSYSSTLEGQAAKEFIAWLV | 1, 3, 4, 5, 6, 10, 11, 14 |
| Liraglutide | HAEGTFTSDVSSYLEGQAAKEEFIAWLVRG | 1, 2, 3, 10, 13, 14 |
| Nesiritide | SPKMVQGSGCFGRKMDRISSSSGLGCKVLR | 4, 5, 6, 15, 19, 29, 32 |
5.2. Target Receptor Structures
| Receptor | PDB ID |
Binding Site (Position of Residues) |
Note |
| Therapeutic Peptide Role: Antagonist | |||
| Angiotensin II Type 1 Receptor (AT1R) | 4YAY [51] | 35, 84, 88, 105, 108, 109, 112, 163, 167, 182, 288, 292 | To reduce vasoconstriction and blood pressure |
| Beta-adrenergic Receptor (β1AR) | 7BTS [52] | 884, 899, 940, 942, 943, 955, 966, 970, 1004, 1005, 1006, 1007, 1009, 1015, 1022, 1032, 1033, 1034, 1035, 1038, 1057, 1121, 1123, 1138, 1209, 1212, 1215, 1218, 1221, 1222, 1228, 1232, 1234, 1321, 1340, 1344, 1349, 1350, 1363, 1364, 1379, 1384 | To reduce heart rate and workload on the hear |
| Interleukin-6 Receptor (IL-6R) | 1N26 [53] | 46, 69, 72, 90, 91, 92, 122, 123, 124 | To reduce inflammation and immune response |
| Mineralocorticoid Receptor (MR) | 1Y9R [54] | 769, 770, 772, 773, 776, 807, 810, 814, 817, 845, 941, 942, 945, 954 | To prevent sodium retention and reduce blood pressure |
| Neuropeptide Y Receptor Y1 (NPY1R) | 7VGX [55] | 117, 120, 121, 124, 173, 200, 212, 215, 219, 220, 280, 282, 283, 284, 286, 287, 294, 298, 299, 302 | To reduce vasoconstriction and sympathetic nervous system activity |
| Platelet-Derived Growth Factor Receptor Alpha (PDGFR) | 6JOL [56] | 625, 627, 644, 648, 658, 674, 676, 677, 814, 815, 816, 825, 835, 836, 837 | To inhibit smooth muscle proliferation and plaque formation |
| Renin Receptor (ATP6AP2) | 3LC8 [57] | 82, 155, 210, 230, 287, 306, 310, 363, 366, 367, 368, 370 | To inhibit renin activity and reduce blood pressure |
| Sphingosine-1-Phosphate Receptor 1 (S1PR1) | 7TD3 [58] | 29, 34, 46, 53, 98, 110, 121, 175, 210, 276, 297 | To inhibit smooth muscle proliferation and vascular inflammation |
| Thrombopoietin Receptor (TPO-R) | 8G04 [59] | 288, 290, 291, 292, 300, 302, 303, 304, 349, 390, 473, 475, 476, 477 | To reduce platelet activation and prevent thrombosis |
| Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) | 4ASD [60] | 840, 848, 866, 868, 885, 889, 899, 916, 917, 918, 919, 922, 1019, 1026, 1035, 1044, 1045, 1046, 1047 | To inhibit angiogenesis and reduce plaque formation |
| Therapeutic Peptide Role: Agonist | |||
| Adenosine A2A Receptor | 8FYN [61] | 85, 168, 169, 177, 246, 249, 250, 253, 264, 267, 270, 274 | To promote vasodilation and reduce inflammation |
| Apelin Receptor (APJ) | 5VBL [30] | 20, 21, 22, 23, 110, 114, 160, 163, 164, 175, 183, 198, 201, 202, 268, 271, 291, 1006, 1009, 1039, 1042 | To improve cardiovascular function |
| Calcium-Sensing Receptor (CaSR) | 7M3F [62] | 20, 21, 22, 23, 24, 41, 54, 57, 59, 60, 98, 101, 106, 109, 112, 116, 117, 119, 132, 350 | To promote vasodilation and calcium homeostasis |
| Glucagon-Like Peptide-1 Receptor (GLP-1R) | 7RTB [63] | 29, 42, 65, 75, 85, 96, 120, 137, 144, 152, 190, 197, 214, 233, 298, 372, 391 | To improve glucose metabolism and reduce CAD risk |
| Low-Density Lipoprotein Receptor (LDLR) | 2FCW [64] | 86, 87, 88, 89, 90, 98, 105, 108, 110, 112, 118, 119, 129, 133, 135, 137, 142, 144, 147, 149, 151, 153, 157, 158 | To enhance LDL clearance and reduce cholesterol levels |
5.3. Computational Tools
5.4. Computing Power
- Sequence Preparation: The first step in the structure modeling pipeline involved formatting the amino acid sequences of the therapeutic peptides according to the requirements of each structure prediction tool. The sequences were thoroughly checked for completeness and alignment with available experimental data.
- Prediction with AlphaFold 3: The next step in the pipeline involved using AlphaFold 3, a deep learning-based tool that predicts high-resolution atomic structures. Peptide sequences were input into AlphaFold 3's web interface, where the tool utilized deep neural networks to generate 3D structures. The accuracy of the predicted structures was assessed based on per-residue confidence scores (pLDDT), which indicate how reliable the predictions are.
- Modeling with I-TASSER: The sequences were also modeled using I-TASSER, a tool that combines template-based modeling with ab initio simulations for additional structural insights. Peptide sequences were aligned against a template library to identify structurally similar proteins. The tool's threading algorithms generated initial models, which were then refined through iterative ab initio simulations. The quality of the resulting models was assessed using the C-score and TM-score, with higher values indicating better model quality.
- De Novo Prediction with PEP-FOLD 4: To complement the results from AlphaFold 3 and I-TASSER, PEP-FOLD 4 was used for de novo prediction of peptide structures. PEP-FOLD 4 generated conformational ensembles for each peptide using fragment libraries and performing energy minimization.
- Structural Optimization: All models underwent a structural optimization process once the initial structures were predicted. This involved energy minimization using molecular mechanics force fields such as AMBER or OPLS to remove steric clashes and improve the overall stability of the structures.
- Validation of Structures: Several validation tools were used to ensure the quality and reliability of the predicted structures. Stereochemical analysis was conducted using Ramachandran plots generated by PROCHECK [75] to assess the backbone dihedral angles. Additionally, MolProbity [76] was used to identify potential structural outliers and errors in the predicted models. Any structures showing significant deviations were refined using visualization and editing tools such as PyMOL [77] and Chimera [78].
- Comparison Across Tools: Finally, the structures predicted by AlphaFold 3, I-TASSER, and PEP-FOLD 4 were compared to identify the most reliable confirmations. Key parameters such as secondary structure elements, folding patterns, and overall energy were analyzed. Based on these comparisons, the best model for each therapeutic peptide was then selected for use in the subsequent molecular docking simulations.
5.6. Peptide-Protein Docking Simulation
5.7. Molecular Dynamics (MD) Simulation
5.8. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
5.9. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Młynarska, E.; Czarnik, W.; Fularski, P.; Hajdys, J.; Majchrowicz, G.; Stabrawa, M.; Rysz, J.; Franczyk, B. , From Atherosclerotic Plaque to Myocardial Infarction—The Leading Cause of Coronary Artery Occlusion. Int. J. Mol. Sci. 2024, 25, 1–27. [Google Scholar] [CrossRef]
- Andreassi, M. G. , Coronary atherosclerosis and somatic mutations: an overview of the contributive factors for oxidative DNA damage. Mutat. Res. /Rev. Mutat. Res. 2003, 543, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Hiwale, K. , Updates in the Management of Coronary Artery Disease: A Review Article. Cureus 2023, 15, e50644. [Google Scholar] [CrossRef]
- Kodeboina, M.; Piayda, K.; Jenniskens, I.; Vyas, P.; Chen, S.; Pesigan, R. J.; Ferko, N.; Patel, B. P.; Dobrin, A.; Habib, J.; Franke, J. , Challenges and Burdens in the Coronary Artery Disease Care Pathway for Patients Undergoing Percutaneous Coronary Intervention: A Contemporary Narrative Review. Int J Env. Res Public Health 2023, 20. [Google Scholar] [CrossRef]
- Libby, P.; Theroux, P. , Pathophysiology of Coronary Artery Disease. Circulation 2005, 111, 3481–3488. [Google Scholar] [CrossRef]
- Rossino, G.; Marchese, E.; Galli, G.; Verde, F.; Finizio, M.; Serra, M.; Linciano, P.; Collina, S. , Peptides as Therapeutic Agents: Challenges and Opportunities in the Green Transition Era. Molecules 2023, 28. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. , Therapeutic peptides: current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Aslan, A.; Ari Yuka, S. , Therapeutic peptides for coronary artery diseases: in silico methods and current perspectives. Amino Acids 2024, 56, 37. [Google Scholar] [CrossRef]
- Alotaiq, N.; Dermawan, D.; Elwali, N. E. , Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 10818. [Google Scholar] [CrossRef]
- Müller, T. D.; Finan, B.; Bloom, S. R.; D'Alessio, D.; Drucker, D. J.; Flatt, P. R.; Fritsche, A.; Gribble, F.; Grill, H. J.; Habener, J. F.; Holst, J. J.; Langhans, W.; Meier, J. J.; Nauck, M. A.; Perez-Tilve, D.; Pocai, A.; Reimann, F.; Sandoval, D. A.; Schwartz, T. W.; Seeley, R. J.; Stemmer, K.; Tang-Christensen, M.; Woods, S. C.; DiMarchi, R. D.; Tschöp, M. H. , Glucagon-like peptide 1 (GLP-1). Mol Metab 2019, 30, 72–130. [Google Scholar]
- Fernandez Rico, C.; Konate, K.; Josse, E.; Nargeot, J.; Barrère-Lemaire, S.; Boisguérin, P. , Therapeutic Peptides to Treat Myocardial Ischemia-Reperfusion Injury. Front Cardiovasc Med 2022, 9, 792885. [Google Scholar] [CrossRef] [PubMed]
- Kazmirchuk, T. D. D.; Bradbury-Jost, C.; Withey, T. A.; Gessese, T.; Azad, T.; Samanfar, B.; Dehne, F.; Golshani, A. , Peptides of a Feather: How Computation Is Taking Peptide Therapeutics under Its Wing. Genes 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Charih, F.; Biggar, K. K.; Green, J. R. , Assessing sequence-based protein-protein interaction predictors for use in therapeutic peptide engineering. Sci Rep 2022, 12, 9610. [Google Scholar] [CrossRef]
- Musliha, A.; Dermawan, D.; Rahayu, P.; Tjandrawinata, R. R. , Unraveling modulation effects on albumin synthesis and inflammation by Striatin, a bioactive protein fraction isolated from Channa striata: In silico proteomics and in vitro approaches. Heliyon 2024, 10, e38386. [Google Scholar] [CrossRef] [PubMed]
- Rahayu, P.; Dermawan, D.; Nailufar, F.; Sulistyaningrum, E.; Tjandrawinata, R. R. , Unlocking the wound-healing potential: An integrative in silico proteomics and in vivo analysis of Tacorin, a bioactive protein fraction from Ananas comosus (L.) Merr. Stem. Biochim. Et Biophys. Acta (BBA) - Proteins Proteom. 2025, 1873, 141060. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, X.; Wei, Z.; Lu, H.; Liu, H. , Reinforcement learning-driven exploration of peptide space: accelerating generation of drug-like peptides. Brief. Bioinform. 2024, 25. [Google Scholar] [CrossRef] [PubMed]
- Motmaen, A.; Dauparas, J.; Baek, M.; Abedi, M.; Baker, D.; Bradley, P. , Peptide-binding specificity prediction using fine-tuned protein structure prediction networks. Proc. Natl. Acad. Sci. United States Am. 2023, 120, e2216697120. [Google Scholar] [CrossRef]
- Saini, R. S.; Binduhayyim, R. I. H.; Gurumurthy, V.; Alshadidi, A. A. F.; Aldosari, L. I. N.; Okshah, A.; Kuruniyan, M. S.; Dermawan, D.; Avetisyan, A.; Mosaddad, S. A.; Heboyan, A. , Dental biomaterials redefined: molecular docking and dynamics-driven dental resin composite optimization. BMC Oral Health 2024, 24, 557. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. , Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–50. [Google Scholar] [CrossRef] [PubMed]
- Wodak, S. J.; Vajda, S.; Lensink, M. F.; Kozakov, D.; Bates, P. A. , Critical Assessment of Methods for Predicting the 3D Structure of Proteins and Protein Complexes. Annu Rev Biophys 2023, 52, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Alotaiq, N.; Dermawan, D. , Advancements in Virtual Bioequivalence: A Systematic Review of Computational Methods and Regulatory Perspectives in the Pharmaceutical Industry. Pharmaceutics 2024, 16, 1414. [Google Scholar] [CrossRef]
- Chu, L. S.; Ruffolo, J. A.; Harmalkar, A.; Gray, J. J. , Flexible protein-protein docking with a multitrack iterative transformer. Protein Sci 2024, 33, e4862. [Google Scholar] [CrossRef]
- Hou, T.; Qiao, X.; Zhang, W.; Xu, X. , Empirical Aqueous Solvation Models Based on Accessible Surface Areas with Implicit Electrostatics. J. Phys. Chem. B 2002, 106, 11295–11304. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S. A. A.; Ballard, A. J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A. W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D. , Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Chen, F.; Kang, Y.; Sun, H.; Liu, H.; Li, D.; Zhu, F.; Hou, T. , HawkRank: a new scoring function for protein-protein docking based on weighted energy terms. J Cheminform 2017, 9, 66. [Google Scholar] [CrossRef]
- Mughal, A.; O'Rourke, S. T. , Vascular effects of apelin: Mechanisms and therapeutic potential. Pharmacol Ther 2018, 190, 139–147. [Google Scholar] [CrossRef]
- Chapman, F. A.; Maguire, J. J.; Newby, D. E.; Davenport, A. P.; Dhaun, N. , Targeting the apelin system for the treatment of cardiovascular diseases. Cardiovasc Res 2023, 119, 2683–2696. [Google Scholar] [CrossRef]
- Liu, W.; Yan, J.; Pan, W.; Tang, M. , Apelin/Elabela-APJ: a novel therapeutic target in the cardiovascular system. Ann. Transl. Med. 2020, 8, 243. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C. U.; Hilberg, O.; Mellemkjær, S.; Nielsen-Kudsk, J. E.; Simonsen, U. , Apelin and pulmonary hypertension. Pulm Circ 2011, 1, 334–46. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yue, Y.; Ma, Y.; Zhang, Q.; Zhou, Q.; Song, Y.; Shen, Y.; Li, X.; Ma, X.; Li, C.; Hanson, M. A.; Han, G. W.; Sickmier, E. A.; Swaminath, G.; Zhao, S.; Stevens, R. C.; Hu, L. A.; Zhong, W.; Zhang, M.; Xu, F. , Structural Basis for Apelin Control of the Human Apelin Receptor. Structure 2017, 25, 858–866.e4. [Google Scholar] [CrossRef]
- Narayanan, S.; Harris, D. L.; Maitra, R.; Runyon, S. P. , Regulation of the Apelinergic System and Its Potential in Cardiovascular Disease: Peptides and Small Molecules as Tools for Discovery. J Med Chem 2015, 58, 7913–27. [Google Scholar] [CrossRef] [PubMed]
- Dermawan, D.; Alotaiq, N. Targeting Cardiovascular Disease Receptors with Antimicrobial Peptides (AMPs): Molecular Docking and Dynamics Insights. In Preprints, Preprints: 2024.
- Xin, X.-Y.; Ruan, C.-H.; Liu, Y.-H.; Jin, H.-N.; Park, S.-K.; Hur, S.-J.; Li, X.-Z.; Choi, S.-H. , Identification of novel antioxidant and anti-inflammatory peptides from bovine hemoglobin by computer simulation of enzymolysis, molecular docking and molecular dynamics. Curr. Res. Food Sci. 2024, 9, 100931. [Google Scholar] [CrossRef]
- Wang, D. D.; Ou-Yang, L.; Xie, H.; Zhu, M.; Yan, H. , Predicting the impacts of mutations on protein-ligand binding affinity based on molecular dynamics simulations and machine learning methods. Comput Struct Biotechnol J 2020, 18, 439–454. [Google Scholar] [CrossRef]
- Williams, M.; Ladbury, J. , Hydrogen Bonds in Protein-Ligand Complexes. 2005; pp 137-161.
- Buse, J. B.; Rosenstock, J.; Sesti, G.; Schmidt, W. E.; Montanya, E.; Brett, J. H.; Zychma, M.; Blonde, L. , Liraglutide once a day versus exenatide twice a day for type 2 diabetes: a 26-week randomised, parallel-group, multinational, open-label trial (LEAD-6). Lancet 2009, 374, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Buse, J. B.; Sesti, G.; Schmidt, W. E.; Montanya, E.; Chang, C. T.; Xu, Y.; Blonde, L.; Rosenstock, J. , Switching to once-daily liraglutide from twice-daily exenatide further improves glycemic control in patients with type 2 diabetes using oral agents. Diabetes Care 2010, 33, 1300–3. [Google Scholar] [CrossRef] [PubMed]
- Rossin, D.; Vanni, R.; Lo Iacono, M.; Cristallini, C.; Giachino, C.; Rastaldo, R. , APJ as Promising Therapeutic Target of Peptide Analogues in Myocardial Infarction- and Hypertension-Induced Heart Failure. Pharmaceutics 2023, 15. [Google Scholar] [CrossRef]
- Zhong, J.-C.; Zhang, Z.-Z.; Wang, W.; McKinnie, S. M. K.; Vederas, J. C.; Oudit, G. Y. , Targeting the apelin pathway as a novel therapeutic approach for cardiovascular diseases. Biochim. Et Biophys. Acta (BBA) - Mol. Basis Dis. 2017, 1863, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Chen, H. , Therapeutic potential of apelin and Elabela in cardiovascular disease. Biomed. Pharmacother. 2023, 166, 115268. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.-C.; Yu, X.-Y.; Huang, Y.; Yung, L.-M.; Lau, C.-W.; Lin, S.-G. , Apelin modulates aortic vascular tone via endothelial nitric oxide synthase phosphorylation pathway in diabetic mice. Cardiovasc. Res. 2007, 74, 388–395. [Google Scholar] [CrossRef]
- Ababei, D. C.; Bild, V.; Macadan, I.; Vasincu, A.; Rusu, R. N.; Blaj, M.; Stanciu, G. D.; Lefter, R. M.; Bild, W. , Therapeutic Implications of Renin-Angiotensin System Modulators in Alzheimer's Dementia. Pharmaceutics 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Koumallos, N.; Sigala, E.; Milas, T.; Baikoussis, N. G.; Aragiannis, D.; Sideris, S.; Tsioufis, K. , Angiotensin Regulation of Vascular Homeostasis: Exploring the Role of ROS and RAS Blockers. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. C.; Wang, Z. , Precision Medicine: Disease Subtyping and Tailored Treatment. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Yin, S.; Mi, X.; Shukla, D. , Leveraging Machine Learning Models for Peptide-Protein Interaction Prediction. ArXiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Nissan, N.; Allen, M. C.; Sabatino, D.; Biggar, K. K. , Future Perspective: Harnessing the Power of Artificial Intelligence in the Generation of New Peptide Drugs. Biomolecules 2024, 14. [Google Scholar] [CrossRef]
- Hashemi, S.; Vosough, P.; Taghizadeh, S.; Savardashtaki, A. , Therapeutic peptide development revolutionized: Harnessing the power of artificial intelligence for drug discovery. Heliyon 2024, 10, e40265. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef]
- Chen, X.; Ji, Z. L.; Chen, Y. Z. , TTD: Therapeutic Target Database. Nucleic Acids Res 2002, 30, 412–5. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. , CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 2018, 46, 363–367. [Google Scholar] [CrossRef]
- Zhang, H.; Unal, H.; Gati, C.; Han, Gye W. ; Liu, W.; Zatsepin, Nadia A.; James, D.; Wang, D.; Nelson, G.; Weierstall, U.; Sawaya, Michael R.; Xu, Q.; Messerschmidt, M.; Williams, Garth J.; Boutet, S.; Yefanov, Oleksandr M.; White, Thomas A.; Wang, C.; Ishchenko, A.; Tirupula, Kalyan C.; Desnoyer, R.; Coe, J.; Conrad, Chelsie E.; Fromme, P.; Stevens, Raymond C.; Katritch, V.; Karnik, Sadashiva S.; Cherezov, V., Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography. Cell 2015, 161, 833–844. [Google Scholar] [PubMed]
- Xu, X.; Kaindl, J.; Clark, M. J.; Hübner, H.; Hirata, K.; Sunahara, R. K.; Gmeiner, P.; Kobilka, B. K.; Liu, X. , Binding pathway determines norepinephrine selectivity for the human β1AR over β2AR. Cell Res. 2021, 31, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J. N.; Moritz, R. L.; Lou, M. Z.; van Donkelaar, A.; Ji, H.; Ivancic, N.; Branson, K. M.; Hall, N. E.; Simpson, R. J. , Structure of the extracellular domains of the human interleukin-6 receptor α-chain. Proc. Natl. Acad. Sci. 2002, 99, 15959–15964. [Google Scholar] [CrossRef]
- Fagart, J.; Huyet, J.; Pinon, G. M.; Rochel, M.; Mayer, C.; Rafestin-Oblin, M.-E. , Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension. Nat. Struct. Mol. Biol. 2005, 12, 554–555. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, J.; Ko, S.-B.; Choi, Y. K.; Jeong, H.; Woo, H.; Kang, H.; Bang, I.; Kim, S. A.; Yoon, T.-Y.; Seok, C.; Im, W.; Choi, H.-J. , Structural basis of neuropeptide Y signaling through Y1 receptor. Nat. Commun. 2022, 13, 853. [Google Scholar] [CrossRef] [PubMed]
- Madej, T.; Lanczycki, C. J.; Zhang, D.; Thiessen, P. A.; Geer, R. C.; Marchler-Bauer, A.; Bryant, S. H. , MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res 2014, (Database issue), D297–303. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, X.; Michael Garavito, R. , Structural analysis of the intracellular domain of (pro)renin receptor fused to maltose-binding protein. Biochem. Biophys. Res. Commun. 2011, 407, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Paknejad, N.; Zhu, L.; Kihara, Y.; Ray, M.; Chun, J.; Liu, W.; Hite, R. K.; Huang, X.-Y. , Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate. Nat. Commun. 2022, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, N.; Masoumi, Z.; James, S. C.; Tucker, J. A.; Winkelmann, H.; Grey, W.; Picton, L. K.; Moss, L.; Wilson, S. C.; Caveney, N. A.; Jude, K. M.; Gati, C.; Piehler, J.; Hitchcock, I. S.; Garcia, K. C. , Structure of the thrombopoietin-MPL receptor complex is a blueprint for biasing hematopoiesis. Cell 2023, 186, 4189–4203.e22. [Google Scholar] [CrossRef] [PubMed]
- McTigue, M.; Murray, B. W.; Chen, J. H.; Deng, Y.-L.; Solowiej, J.; Kania, R. S. , Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc. Natl. Acad. Sci. 2012, 109, 18281–18289. [Google Scholar] [CrossRef]
- Martynowycz, M. W.; Shiriaeva, A.; Clabbers, M. T. B.; Nicolas, W. J.; Weaver, S. J.; Hattne, J.; Gonen, T. , A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals. Nat. Commun. 2023, 14, 1086. [Google Scholar] [CrossRef]
- Gao, Y.; Robertson, M. J.; Rahman, S. N.; Seven, A. B.; Zhang, C.; Meyerowitz, J. G.; Panova, O.; Hannan, F. M.; Thakker, R. V.; Bräuner-Osborne, H.; Mathiesen, J. M.; Skiniotis, G. , Asymmetric activation of the calcium-sensing receptor homodimer. Nature 2021, 595, 455–459. [Google Scholar] [CrossRef]
- Johnson, R. M.; Zhang, X.; Piper, S. J.; Nettleton, T. J.; Vandekolk, T. H.; Langmead, C. J.; Danev, R.; Sexton, P. M.; Wootten, D. , Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor. Biochem. Biophys. Res. Commun. 2021, 578, 84–90. [Google Scholar] [CrossRef]
- Fisher, C.; Beglova, N.; Blacklow, S. C. , Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors. Mol. Cell 2006, 22, 277–283. [Google Scholar] [CrossRef]
- Desai, D.; Kantliwala, S. V.; Vybhavi, J.; Ravi, R.; Patel, H.; Patel, J. , Review of AlphaFold 3: Transformative Advances in Drug Design and Therapeutics. Cureus 2024, 16, e63646. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. , Protein Structure and Function Prediction Using I-TASSER. 2015; Vol. 52, pp 5.8.1-5.8.15.
- Zhang, Y. , Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins 2014, 82. [Google Scholar] [CrossRef] [PubMed]
- Rey, J.; Murail, S.; de Vries, S.; Derreumaux, P.; Tuffery, P. , PEP-FOLD4: a pH-dependent force field for peptide structure prediction in aqueous solution. Nucleic Acids Res 2023, 51, W432–w437. [Google Scholar] [CrossRef] [PubMed]
- Maupetit, J.; Tuffery, P. , PEP-FOLD: An online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009, 37, W498–W503. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Maupetit, J.; Tuffery, P. , Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Honorato, R.; Trellet, M.; Jimenez-Garcia, B.; Schaarschmidt, J.; Giulini, M.; Reys, V.; Koukos, P.; Rodrigues, J.; Karaca, E.; Zundert, G.; Roel-Touris, J.; van Noort, C.; Jandova, Z.; Melquiond, A.; Bonvin, A. , The HADDOCK2.4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jin, B.; Li, H.; Huang, S. Y. , HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res 2018, 46, W443–w450. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D. R.; Xia, B.; Porter, K. A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. , The ClusPro web server for protein-protein docking. Nat Protoc 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Weng, G.; Wang, E.; Wang, Z.; Liu, H.; Zhu, F.; Li, D.; Hou, T. , HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res 2019, 47, W322–w330. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R. A.; MacArthur, M. W.; Moss, D. S.; Thornton, J. M. , PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Williams, C. J.; Headd, J. J.; Moriarty, N. W.; Prisant, M. G.; Videau, L. L.; Deis, L. N.; Verma, V.; Keedy, D. A.; Hintze, B. J.; Chen, V. B.; Jain, S.; Lewis, S. M.; Arendall, W. B., 3rd; Snoeyink, J.; Adams, P. D.; Lovell, S. C.; Richardson, J. S.; Richardson, D. C. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Schrödinger<i> The PyMOL Molecular Graphics System</i>, 2. Schrödinger The PyMOL Molecular Graphics System, 2.4; 2020.
- Pettersen, E. F.; Goddard, T. D.; Huang, C. C.; Couch, G. S.; Greenblatt, D. M.; Meng, E. C.; Ferrin, T. E. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Johansson, M.; Zoete, V.; Michielin, O.; Guex, N. , Defining and searching for structural motifs using DeepView/Swiss-PDBViewer. BMC Bioinform. 2012, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Xue, L. C.; Rodrigues, J. P.; Kastritis, P. L.; Bonvin, A. M.; Vangone, A. , PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Bonvin, A. , PRODIGY: A Contact-based Predictor of Binding Affinity in Protein-protein Complexes. BIO-PROTOCOL.
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M. R.; Smith, J. C.; Kasson, P. M.; van der Spoel, D.; Hess, B.; Lindahl, E. , GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Robertson, M. J.; Tirado-Rives, J.; Jorgensen, W. L. , Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J Chem Theory Comput 2015, 11, 3499–509. [Google Scholar] [CrossRef]
- Yuet, P.; Blankschtein, D. , Molecular Dynamics Simulation Study of Water Surfaces: Comparison of Flexible Water Models. J. Phys. Chem.. B 2010, 114, 13786–13795. [Google Scholar] [CrossRef] [PubMed]
- Doni Dermawan, F. A. , Nasr Eldin Elwali, Nasser Alotaiq, Therapeutic potential of earthworm-derived proteins: Targeting NEDD4 for cardiovascular disease intervention. Issue: 1: 2024; Vol. Volume: 15, p 216-232.
- Tian, S.; Sun, H.; Pan, P.; Li, D.; Zhen, X.; Li, Y.; Hou, T. , Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model 2014, 54, 2664–2679. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M. S.; Valdés-Tresanco, M. E.; Valiente, P. A.; Moreno, E. , gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Miller, B. R., 3rd; McGee, T. D., Jr.; Swails, J. M.; Homeyer, N.; Gohlke, H.; Roitberg, A. E. , MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Panday, S. K.; Alexov, E. , Protein-Protein Binding Free Energy Predictions with the MM/PBSA Approach Complemented with the Gaussian-Based Method for Entropy Estimation. ACS Omega 2022, 7, 11057–11067. [Google Scholar] [CrossRef] [PubMed]
- OriginLab Origin(Pro), 2022; OriginLab Corporation: Northampton, MA, USA, 2022.
 |
| Receptor | Therapeutic Peptide | Docking Platform | Binding Affinity (kcal/mol) |
|---|---|---|---|
| AT1R | Apelin | ClusPro 2.0 | -15.6 |
| β1AR | Apelin | ClusPro 2.0 | -17.8 |
| IL-6R | Apelin | ClusPro 2.0 | -19.1 |
| MR | Liraglutide | HawkDock 2.0 | -11.0 |
| NPY1R | ANP | HawkDock 2.0 | -20.1 |
| PDGFR | Apelin | ClusPro 2.0 | -13.8 |
| ATP6AP2 | Apelin | HawkDock 2.0 | -16.2 |
| S1PR1 | ANP | HADDOCK 2.4 | -10.0 |
| TPO-R | FX06 | ClusPro 2.0 | -12.2 |
| VEGFR2 | Apelin | HawkDock 2.0 | -21.7 |
| Receptor | Therapeutic Peptide | Docking Platform | Binding Affinity (kcal/mol) |
| A2A | ANP | HawkDock 2.0 | -12.7 |
| APJ | Apelin | ClusPro 2.0 | -12.3 |
| CaSR | Exenatide | HADDOCK 2.4 | -13.2 |
| GLP-1R | Apelin | HawkDock 2.0 | -13.4 |
| LDLR | ANP | HPEPDOCK 2.0 | -11.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





