Submitted:
26 August 2023
Posted:
29 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Why Antisense Technology?
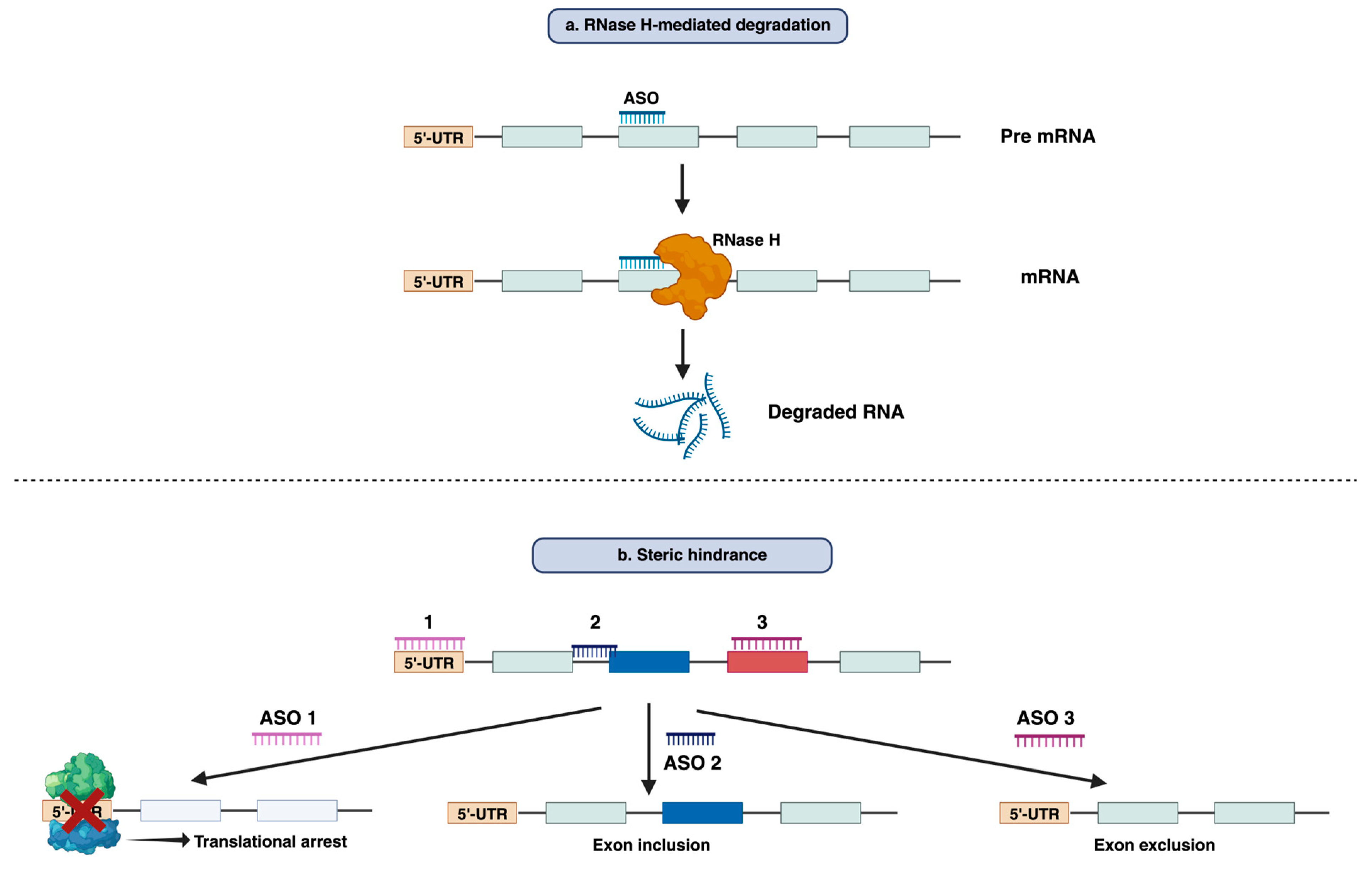
3. The mode of action of Antisense Oligonucleotide
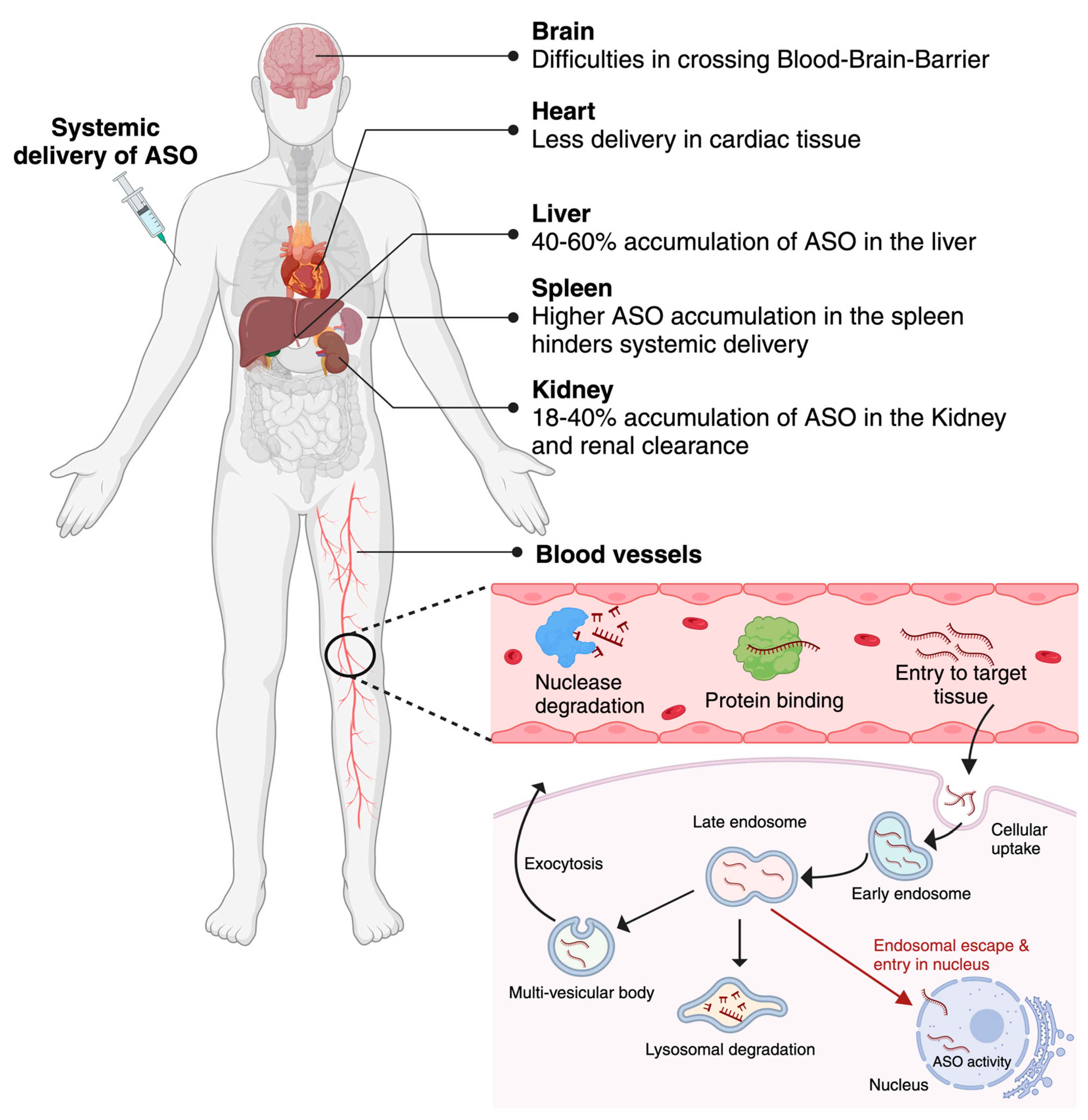
4. Molecular mechanism of cellular uptake and intracellular distribution of Antisense Oligonucleotides
5. Challenges associated with ASOs delivery
6. Strategies to enhance the stability and delivery of Antisense Oligonucleotides
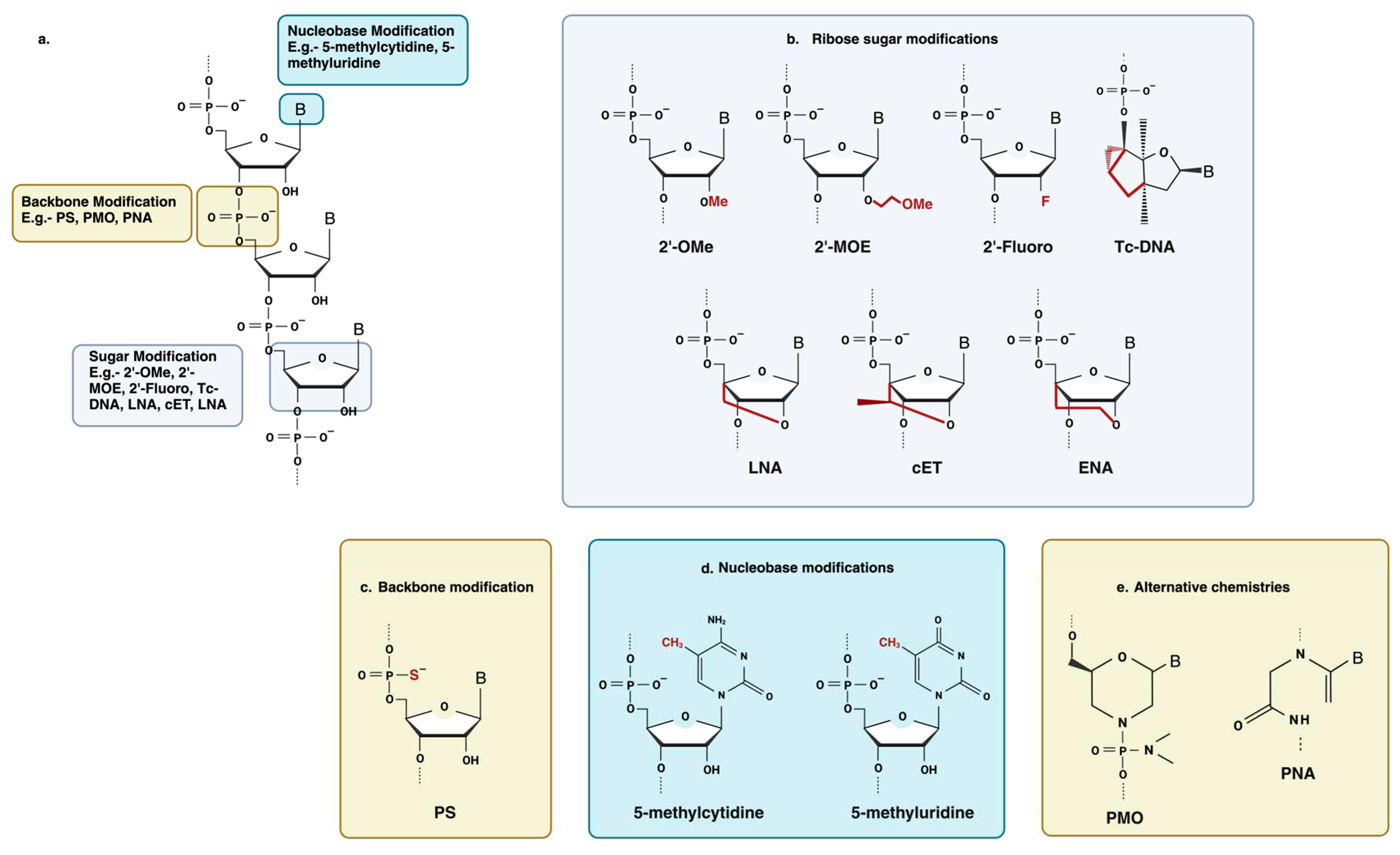
6.1. Chemical modification
6.1.1. Backbone modification
6.1.2. Ribose sugar modification
6.1.3. Nucleobase modification
6.1.4. Other Oligonucleotide Modifications
6.2. Bioconjugates
6.2.1. Cell Penetrating Peptides
7. Overcoming the limitations of PMO by conjugating it with Cell Penetrating Peptides
8. DG9: A CPP for enhancing the delivery and cellular uptake of ASO and proteins
9. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A 1978, 75, 285–288. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A 1978, 75, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Sasso, J.M.; Ambrose, B.J.B.; Tenchov, R.; Datta, R.S.; Basel, M.T.; DeLong, R.K.; Zhou, Q.A. The Progress and Promise of RNA Medicine horizontal line An Arsenal of Targeted Treatments. J Med Chem 2022, 65, 6975–7015. [Google Scholar] [CrossRef] [PubMed]
- Kuijper, E.C.; Bergsma, A.J.; Pijnappel, W.; Aartsma-Rus, A. Opportunities and challenges for antisense oligonucleotide therapies. J Inherit Metab Dis 2021, 44, 72–87. [Google Scholar] [CrossRef]
- Sharma, V.K.; Sharma, R.K.; Singh, S.K. Antisense oligonucleotides: modifications and clinical trials. MedChemComm 2014, 5, 1454–1471. [Google Scholar] [CrossRef]
- Juliano, R.L.; Ming, X.; Carver, K.; Laing, B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther 2014, 24, 101–113. [Google Scholar] [CrossRef]
- Jarver, P.; Coursindel, T.; Andaloussi, S.E.; Godfrey, C.; Wood, M.J.; Gait, M.J. Peptide-mediated Cell and In Vivo Delivery of Antisense Oligonucleotides and siRNA. Mol Ther Nucleic Acids 2012, 1, e27. [Google Scholar] [CrossRef]
- Derossi, D.; Calvet, S.; Trembleau, A.; Brunissen, A.; Chassaing, G.; Prochiantz, A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem 1996, 271, 18188–18193. [Google Scholar] [CrossRef]
- Lim, K.R.Q.; Woo, S.; Melo, D.; Huang, Y.; Dzierlega, K.; Shah, M.N.A.; Aslesh, T.; Roshmi, R.R.; Echigoya, Y.; Maruyama, R.; et al. Development of DG9 peptide-conjugated single- and multi-exon skipping therapies for the treatment of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2022, 119. [Google Scholar] [CrossRef]
- Aslesh, T.; Erkut, E.; Ren, J.; Lim, K.R.Q.; Woo, S.; Hatlevig, S.; Moulton, H.M.; Gosgnach, S.; Greer, J.; Maruyama, R.; et al. DG9-conjugated morpholino rescues phenotype in SMA mice by reaching the CNS via a subcutaneous administration. JCI Insight 2023, 8. [Google Scholar] [CrossRef]
- van Roon-Mom, W.; Ferguson, C.; Aartsma-Rus, A. From Failure to Meet the Clinical Endpoint to U.S. Food and Drug Administration Approval: 15th Antisense Oligonucleotide Therapy Approved Qalsody (Tofersen) for Treatment of SOD1 Mutated Amyotrophic Lateral Sclerosis. Nucleic Acid Ther 2023, 33, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Toonen, L.J.; van Roon-Mom, W.M. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev 2015, 87, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lima, W.F.; Zhang, H.; Fan, A.; Sun, H.; Crooke, S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem 2004, 279, 17181–17189. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Dominski, Z.; Kole, R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A 1993, 90, 8673–8677. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv Neurobiol 2018, 20, 31–61. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Straub, V.; Hemmings, R.; Haas, M.; Schlosser-Weber, G.; Stoyanova-Beninska, V.; Mercuri, E.; Muntoni, F.; Sepodes, B.; Vroom, E.; et al. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther 2017, 27, 251–259. [Google Scholar] [CrossRef]
- Wan, L.; Dreyfuss, G. Splicing-Correcting Therapy for SMA. Cell 2017, 170, 5. [Google Scholar] [CrossRef]
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879. [Google Scholar] [CrossRef]
- Dolgin, E. News Feature: Gene therapy successes point to better therapies. Proc Natl Acad Sci U S A 2019, 116, 23866–23870. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sinhari, A.; Jain, P.; Jadhav, H.R. A perspective on oligonucleotide therapy: Approaches to patient customization. Front Pharmacol 2022, 13, 1006304. [Google Scholar] [CrossRef] [PubMed]
- Migliorati, J.M.; Liu, S.; Liu, A.; Gogate, A.; Nair, S.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X.B. Absorption, Distribution, Metabolism, and Excretion of US Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab Dispos 2022, 50, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L.; Ming, X.; Nakagawa, O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug Chem 2012, 23, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Sun, H.; Nichols, J.G.; Crooke, S.T. RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus. Mol Ther 2017, 25, 2075–2092. [Google Scholar] [CrossRef]
- Echevarria, L.; Aupy, P.; Goyenvalle, A. Exon-skipping advances for Duchenne muscular dystrophy. Hum Mol Genet 2018, 27, R163–R172. [Google Scholar] [CrossRef]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol 2013, 31, 653–658. [Google Scholar] [CrossRef]
- Iversen, F.; Yang, C.; Dagnaes-Hansen, F.; Schaffert, D.H.; Kjems, J.; Gao, S. Optimized siRNA-PEG conjugates for extended blood circulation and reduced urine excretion in mice. Theranostics 2013, 3, 201–209. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 2015, 87, 46–51. [Google Scholar] [CrossRef]
- Tsui, N.B.Y.; Ng, E.K.O.; Lo, Y.M.D. Stability of Endogenous and Added RNA in Blood Specimens, Serum, and Plasma. Clinical Chemistry 2002, 48, 1647–1653. [Google Scholar] [CrossRef]
- Allen, T.M. The use of glycolipids and hydrophilic polymers in avoiding rapid uptake of liposomes by the mononuclear phagocyte system. Advanced Drug Delivery Reviews 1994, 13, 285–309. [Google Scholar] [CrossRef]
- Bijsterbosch, M.K.; Manoharan, M.; Rump, E.T.; De Vrueh, R.L.A.; van Veghel, R.; Tivel, K.L.; Biessen, E.A.L.; Bennett, C.F.; Cook, P.D.; van Berkel, T.J.C. In vivo fate of phosphorothioate antisense oligodeoxynucleotides: predominant uptake by scavenger receptors on endothelial liver cells. Nucleic Acids Research 1997, 25, 3290–3296. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.M. The blood-brain barrier.
- Finkel, R.S.; Chiriboga, C.A.; Vajsar, J.; Day, J.W.; Montes, J.; De Vivo, D.C.; Yamashita, M.; Rigo, F.; Hung, G.; Schneider, E.; et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 2016, 388, 3017–3026. [Google Scholar] [CrossRef] [PubMed]
- Di Fusco, D.; Dinallo, V.; Marafini, I.; Figliuzzi, M.M.; Romano, B.; Monteleone, G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front Pharmacol 2019, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.B.; Seth, P.P. The Medicinal Chemistry of Therapeutic Oligonucleotides. J Med Chem 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther 2014, 24, 374–387. [Google Scholar] [CrossRef]
- Furdon, P.J.; Dominski, Z.; Kole, R. RNase H cleavage of RNA hybridized to oligonucleotides containing methylphosphonate, phosphorothioate and phosphodiester bonds. Nucleic Acids Res 1989, 17, 9193–9204. [Google Scholar] [CrossRef]
- Rinaldi, C.; Wood, M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Ezzat, K.; Aoki, Y.; Koo, T.; McClorey, G.; Benner, L.; Coenen-Stass, A.; O’Donovan, L.; Lehto, T.; Garcia-Guerra, A.; Nordin, J.; et al. Self-Assembly into Nanoparticles Is Essential for Receptor Mediated Uptake of Therapeutic Antisense Oligonucleotides. Nano Lett 2015, 15, 4364–4373. [Google Scholar] [CrossRef]
- Miller, C.M.; Donner, A.J.; Blank, E.E.; Egger, A.W.; Kellar, B.M.; Ostergaard, M.E.; Seth, P.P.; Harris, E.N. Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res 2016, 44, 2782–2794. [Google Scholar] [CrossRef]
- Gaus, H.J.; Gupta, R.; Chappell, A.E.; Ostergaard, M.E.; Swayze, E.E.; Seth, P.P. Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay. Nucleic Acids Res 2019, 47, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.A. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta 1999, 1489, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Kang, S.H.; Gryaznov, S.M.; DeDionisio, L.; Heidenreich, O.; Sullivan, S.; Xu, X.; Nerenberg, M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. Journal of Biological Chemistry 1994, 269, 26801–26805. [Google Scholar] [CrossRef] [PubMed]
- Guvakova, M.A.; Yakubov, L.A.; Vlodavsky, I.; Tonkinson, J.L.; Stein, C.A. Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem 1995, 270, 2620–2627. [Google Scholar] [CrossRef]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Freier, S.M.; Altmann, K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res 1997, 25, 4429–4443. [Google Scholar] [CrossRef]
- Lubini, P.; Zurcher, W.; Egli, M. Stabilizing effects of the RNA 2’-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2’-O-methylated adenosines. Chem Biol 1994, 1, 39–45. [Google Scholar] [CrossRef]
- McKay, R.A.; Miraglia, L.J.; Cummins, L.L.; Owens, S.R.; Sasmor, H.; Dean, N.M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J Biol Chem 1999, 274, 1715–1722. [Google Scholar] [CrossRef]
- Prakash, T.P. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem Biodivers 2011, 8, 1616–1641. [Google Scholar] [CrossRef]
- Frieden, M.; Hansen, H.F.; Koch, T. Nuclease Stability of LNA Oligonucleotides and LNA-DNA Chimeras. Nucleosides, Nucleotides & Nucleic Acids 2003, 22, 1041–1043. [Google Scholar] [CrossRef]
- Braasch, D.A.; Liu, Y.; Corey, D.R. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Research 2002, 30, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Elayadi, A.N.; Braasch, D.A.; Corey, D.R. Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry 2002, 41, 9973–9981. [Google Scholar] [CrossRef]
- Swayze, E.E.; Siwkowski, A.M.; Wancewicz, E.V.; Migawa, M.T.; Wyrzykiewicz, T.K.; Hung, G.; Monia, B.P.; Bennett, C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res 2007, 35, 687–700. [Google Scholar] [CrossRef]
- Gruegelsiepe, H.; Brandt, O.; Hartmann, R.K. Antisense inhibition of RNase P: mechanistic aspects and application to live bacteria. J Biol Chem 2006, 281, 30613–30620. [Google Scholar] [CrossRef] [PubMed]
- Renneberg, D.; Leumann, C.J. Watson-Crick base-pairing properties of tricyclo-DNA. J Am Chem Soc 2002, 124, 5993–6002. [Google Scholar] [CrossRef] [PubMed]
- Renneberg, D.; Bouliong, E.; Reber, U.; Schumperli, D.; Leumann, C.J. Antisense properties of tricyclo-DNA. Nucleic Acids Res 2002, 30, 2751–2757. [Google Scholar] [CrossRef]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Kinberger, G.; Migawa, M.T.; Gaus, H.; Bhat, B.; et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp Ser (Oxf) 2008, 553–554. [Google Scholar] [CrossRef]
- Southwell, A.L.; Skotte, N.H.; Kordasiewicz, H.B.; Ostergaard, M.E.; Watt, A.T.; Carroll, J.B.; Doty, C.N.; Villanueva, E.B.; Petoukhov, E.; Vaid, K.; et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther 2014, 22, 2093–2106. [Google Scholar] [CrossRef]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 2012, 19, 937–954. [Google Scholar] [CrossRef]
- Chenna, V.; Rapireddy, S.; Sahu, B.; Ausin, C.; Pedroso, E.; Ly, D.H. A simple cytosine to G-clamp nucleobase substitution enables chiral gamma-PNAs to invade mixed-sequence double-helical B-form DNA. Chembiochem 2008, 9, 2388–2391. [Google Scholar] [CrossRef]
- Rapireddy, S.; Bahal, R.; Ly, D.H. Strand invasion of mixed-sequence, double-helical B-DNA by gamma-peptide nucleic acids containing G-clamp nucleobases under physiological conditions. Biochemistry 2011, 50, 3913–3918. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchard, O.; Sonnichsen, S.H.; Nielsen, P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Schwarz, F.P.; Robinson, S.; Butler, J.M. Thermodynamic comparison of PNA/DNA and DNA/DNA hybridization reactions at ambient temperature. Nucleic Acids Res 1999, 27, 4792–4800. [Google Scholar] [CrossRef]
- Wittung, P.; Kajanus, J.; Edwards, K.; Nielsen, P.; Norden, B.; Malmstrom, B.G. Phospholipid membrane permeability of peptide nucleic acid. FEBS Lett 1995, 365, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Hyrup, B.; Nielsen, P.E. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem 1996, 4, 5–23. [Google Scholar] [CrossRef]
- McMahon, B.M.; Mays, D.; Lipsky, J.; Stewart, J.A.; Fauq, A.; Richelson, E. Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev 2002, 12, 65–70. [Google Scholar] [CrossRef]
- Summerton, J.; Weller, D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev 1997, 7, 187–195. [Google Scholar] [CrossRef]
- Hudziak, R.M.; Barofsky, E.; Barofsky, D.F.; Weller, D.L.; Huang, S.B.; Weller, D.D. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev 1996, 6, 267–272. [Google Scholar] [CrossRef]
- Lee, J.J.; Yokota, T. Antisense therapy in neurology. J Pers Med 2013, 3, 144–176. [Google Scholar] [CrossRef]
- Sheng, L.; Rigo, F.; Bennett, C.F.; Krainer, A.R.; Hua, Y. Comparison of the efficacy of MOE and PMO modifications of systemic antisense oligonucleotides in a severe SMA mouse model. Nucleic Acids Res 2020, 48, 2853–2865. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Yokota, T. Golodirsen for Duchenne muscular dystrophy. Drugs Today (Barc) 2020, 56, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Roshmi, R.R.; Yokota, T. Viltolarsen for the treatment of Duchenne muscular dystrophy. Drugs Today (Barc) 2019, 55, 627–639. [Google Scholar] [CrossRef]
- Lu, Q.L.; Mann, C.J.; Lou, F.; Bou-Gharios, G.; Morris, G.E.; Xue, S.A.; Fletcher, S.; Partridge, T.A.; Wilton, S.D. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med 2003, 9, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Gebski, B.L.; Mann, C.J.; Fletcher, S.; Wilton, S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet 2003, 12, 1801–1811. [Google Scholar] [CrossRef]
- Yokota, T.; Lu, Q.L.; Partridge, T.; Kobayashi, M.; Nakamura, A.; Takeda, S.; Hoffman, E. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol 2009, 65, 667–676. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Rigo, F.; Hung, G.; Horev, G.; Bennett, C.F.; Krainer, A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 2011, 478, 123–126. [Google Scholar] [CrossRef]
- Hua, Y.; Liu, Y.H.; Sahashi, K.; Rigo, F.; Bennett, C.F.; Krainer, A.R. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev 2015, 29, 288–297. [Google Scholar] [CrossRef]
- Porensky, P.N.; Mitrpant, C.; McGovern, V.L.; Bevan, A.K.; Foust, K.D.; Kaspar, B.K.; Wilton, S.D.; Burghes, A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet 2012, 21, 1625–1638. [Google Scholar] [CrossRef]
- Zhou, H.; Janghra, N.; Mitrpant, C.; Dickinson, R.L.; Anthony, K.; Price, L.; Eperon, I.C.; Wilton, S.D.; Morgan, J.; Muntoni, F. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum Gene Ther 2013, 24, 331–342. [Google Scholar] [CrossRef]
- Lehto, T.; Castillo Alvarez, A.; Gauck, S.; Gait, M.J.; Coursindel, T.; Wood, M.J.; Lebleu, B.; Boisguerin, P. Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res 2014, 42, 3207–3217. [Google Scholar] [CrossRef] [PubMed]
- Tsoumpra, M.K.; Fukumoto, S.; Matsumoto, T.; Takeda, S.; Wood, M.J.A.; Aoki, Y. Peptide-conjugate antisense based splice-correction for Duchenne muscular dystrophy and other neuromuscular diseases. EBioMedicine 2019, 45, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Summerton, J.E. Invention and Early History of Morpholinos: From Pipe Dream to Practical Products. Methods Mol Biol 2017, 1565, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Thomas, O.S.; Weber, W. Overcoming Physiological Barriers to Nanoparticle Delivery-Are We There Yet? Front Bioeng Biotechnol 2019, 7, 415. [Google Scholar] [CrossRef] [PubMed]
- Nishina, K.; Unno, T.; Uno, Y.; Kubodera, T.; Kanouchi, T.; Mizusawa, H.; Yokota, T. Efficient In Vivo Delivery of siRNA to the Liver by Conjugation of alpha-Tocopherol. Mol Ther 2008, 16, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 2007, 25, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef]
- Lorenz, C.; Hadwiger, P.; John, M.; Vornlocher, H.P.; Unverzagt, C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett 2004, 14, 4975–4977. [Google Scholar] [CrossRef]
- Klein, A.F.; Varela, M.A.; Arandel, L.; Holland, A.; Naouar, N.; Arzumanov, A.; Seoane, D.; Revillod, L.; Bassez, G.; Ferry, A.; et al. Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. J Clin Invest 2019, 129, 4739–4744. [Google Scholar] [CrossRef]
- Eguchi, A.; Meade, B.R.; Chang, Y.C.; Fredrickson, C.T.; Willert, K.; Puri, N.; Dowdy, S.F. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat Biotechnol 2009, 27, 567–571. [Google Scholar] [CrossRef]
- Betts, C.; Saleh, A.F.; Arzumanov, A.A.; Hammond, S.M.; Godfrey, C.; Coursindel, T.; Gait, M.J.; Wood, M.J. Pip6-PMO, A New Generation of Peptide-oligonucleotide Conjugates With Improved Cardiac Exon Skipping Activity for DMD Treatment. Mol Ther Nucleic Acids 2012, 1, e38. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.R.; Ming, X.; Fisher, M.; Lackey, J.G.; Rajeev, K.G.; Manoharan, M.; Juliano, R.L. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug Chem 2011, 22, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Ammala, C.; Drury, W.J., 3rd; Knerr, L.; Ahlstedt, I.; Stillemark-Billton, P.; Wennberg-Huldt, C.; Andersson, E.M.; Valeur, E.; Jansson-Lofmark, R.; Janzen, D.; et al. Targeted delivery of antisense oligonucleotides to pancreatic beta-cells. Sci Adv 2018, 4, eaat3386. [Google Scholar] [CrossRef]
- Liu, X.; Wang, W.; Samarsky, D.; Liu, L.; Xu, Q.; Zhang, W.; Zhu, G.; Wu, P.; Zuo, X.; Deng, H.; et al. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res 2014, 42, 11805–11817. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O., 2nd; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Zhu, P.; Lee, S.K.; Chowdhury, D.; Kussman, S.; Dykxhoorn, D.M.; Feng, Y.; Palliser, D.; Weiner, D.B.; Shankar, P.; et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol 2005, 23, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Tushir-Singh, J. Antibody-siRNA conjugates: drugging the undruggable for anti-leukemic therapy. Expert Opin Biol Ther 2017, 17, 325–338. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 2014, 136, 16958–16961. [Google Scholar] [CrossRef]
- Matsuda, S.; Keiser, K.; Nair, J.K.; Charisse, K.; Manoharan, R.M.; Kretschmer, P.; Peng, C.G.; A, V.K.i.; Kandasamy, P.; Willoughby, J.L.; et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol 2015, 10, 1181–1187. [Google Scholar] [CrossRef]
- Anwar, S.; Mir, F.; Yokota, T. Enhancing the Effectiveness of Oligonucleotide Therapeutics Using Cell-Penetrating Peptide Conjugation, Chemical Modification, and Carrier-Based Delivery Strategies. Pharmaceutics 2023, 15. [Google Scholar] [CrossRef]
- McClorey, G.; Banerjee, S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Torchilin, V.P. Cell-penetrating peptides: breaking through to the other side. Trends Mol Med 2012, 18, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.H.; Park, J.; Koo, H. Recent advances in selective and targeted drug/gene delivery systems using cell-penetrating peptides. Arch Pharm Res 2023, 46, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front Pharmacol 2020, 11, 697. [Google Scholar] [CrossRef]
- Verdurmen, W.P.; Brock, R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol Sci 2011, 32, 116–124. [Google Scholar] [CrossRef]
- Godfrey, C.; Desviat, L.R.; Smedsrod, B.; Pietri-Rouxel, F.; Denti, M.A.; Disterer, P.; Lorain, S.; Nogales-Gadea, G.; Sardone, V.; Anwar, R.; et al. Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol Med 2017, 9, 545–557. [Google Scholar] [CrossRef]
- Gomez, J.A.; Chen, J.; Ngo, J.; Hajkova, D.; Yeh, I.J.; Gama, V.; Miyagi, M.; Matsuyama, S. Cell-Penetrating Penta-Peptides (CPP5s): Measurement of Cell Entry and Protein-Transduction Activity. Pharmaceuticals (Basel) 2010, 3, 3594–3613. [Google Scholar] [CrossRef]
- Klabenkova, K.; Fokina, A.; Stetsenko, D. Chemistry of Peptide-Oligonucleotide Conjugates: A Review. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Lehto, T.; Ezzat, K.; Wood, M.J.A.; El Andaloussi, S. Peptides for nucleic acid delivery. Adv Drug Deliv Rev 2016, 106, 172–182. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed Pharmacother 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Birnkrant, D.J.; Bushby, K.M.; Amin, R.S.; Bach, J.R.; Benditt, J.O.; Eagle, M.; Finder, J.D.; Kalra, M.S.; Kissel, J.T.; Koumbourlis, A.C.; et al. The respiratory management of patients with duchenne muscular dystrophy: a DMD care considerations working group specialty article. Pediatr Pulmonol 2010, 45, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Manzur, A.Y.; Kinali, M.; Muntoni, F. Update on the management of Duchenne muscular dystrophy. Arch Dis Child 2008, 93, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Wilton-Clark, H.; Yokota, T. Casimersen for Duchenne muscular dystrophy. Drugs Today (Barc) 2021, 57, 707–717. [Google Scholar] [CrossRef]
- Roshmi, R.R.; Yokota, T. Viltolarsen: From Preclinical Studies to FDA Approval. Methods Mol Biol 2023, 2587, 31–41. [Google Scholar] [CrossRef]
- Yin, H.; Moulton, H.M.; Seow, Y.; Boyd, C.; Boutilier, J.; Iverson, P.; Wood, M.J. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum Mol Genet 2008, 17, 3909–3918. [Google Scholar] [CrossRef]
- Wu, B.; Moulton, H.M.; Iversen, P.L.; Jiang, J.; Li, J.; Li, J.; Spurney, C.F.; Sali, A.; Guerron, A.D.; Nagaraju, K.; et al. Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proc Natl Acad Sci U S A 2008, 105, 14814–14819. [Google Scholar] [CrossRef] [PubMed]
- Goyenvalle, A.; Griffith, G.; Babbs, A.; El Andaloussi, S.; Ezzat, K.; Avril, A.; Dugovic, B.; Chaussenot, R.; Ferry, A.; Voit, T.; et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat Med 2015, 21, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Samoylova, T.I.; Smith, B.F. Elucidation of muscle-binding peptides by phage display screening.
- Yin, H.; Moulton, H.M.; Betts, C.; Seow, Y.; Boutilier, J.; Iverson, P.L.; Wood, M.J. A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum Mol Genet 2009, 18, 4405–4414. [Google Scholar] [CrossRef]
- Echigoya, Y.; Nakamura, A.; Nagata, T.; Urasawa, N.; Lim, K.R.Q.; Trieu, N.; Panesar, D.; Kuraoka, M.; Moulton, H.M.; Saito, T.; et al. Effects of systemic multiexon skipping with peptide-conjugated morpholinos in the heart of a dog model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2017, 114, 4213–4218. [Google Scholar] [CrossRef]
- Therapeutics, P.E.O. Available online: https://investors.pepgen.com/static-files/7244e66d-1227-42b1-841fbf95306d84b1.
- Abes, S.; Turner, J.J.; Ivanova, G.D.; Owen, D.; Williams, D.; Arzumanov, A.; Clair, P.; Gait, M.J.; Lebleu, B. Efficient splicing correction by PNA conjugation to an R6-Penetratin delivery peptide. Nucleic Acids Res 2007, 35, 4495–4502. [Google Scholar] [CrossRef]
- Bersani, M.; Rizzuti, M.; Pagliari, E.; Garbellini, M.; Saccomanno, D.; Moulton, H.M.; Bresolin, N.; Comi, G.P.; Corti, S.; Nizzardo, M. Cell-penetrating peptide-conjugated Morpholino rescues SMA in a symptomatic preclinical model. Mol Ther 2022, 30, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. Journal of Biological Chemistry 1994, 269, 10444–10450. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Joliot, A.; Bloch-Gallego, E.; Zahraoui, A.; Triller, A.; Prochiantz, A. Antennapedia homeobox as a signal for the cellular internalization and nuclear addressing of a small exogenous peptide. J Cell Sci 1992, 102 Pt 4, 717–722. [Google Scholar] [CrossRef]
- Yin, H.; Saleh, A.F.; Betts, C.; Camelliti, P.; Seow, Y.; Ashraf, S.; Arzumanov, A.; Hammond, S.; Merritt, T.; Gait, M.J.; et al. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol Ther 2011, 19, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, C.; Muses, S.; McClorey, G.; Wells, K.E.; Coursindel, T.; Terry, R.L.; Betts, C.; Hammond, S.; O’Donovan, L.; Hildyard, J.; et al. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. Hum Mol Genet 2015, 24, 4225–4237. [Google Scholar] [CrossRef] [PubMed]
- Betts, C.A.; Saleh, A.F.; Carr, C.A.; Hammond, S.M.; Coenen-Stass, A.M.; Godfrey, C.; McClorey, G.; Varela, M.A.; Roberts, T.C.; Clarke, K.; et al. Prevention of exercised induced cardiomyopathy following Pip-PMO treatment in dystrophic mdx mice. Sci Rep 2015, 5, 8986. [Google Scholar] [CrossRef]
- Ivanova, G.D.; Arzumanov, A.; Abes, R.; Yin, H.; Wood, M.J.; Lebleu, B.; Gait, M.J. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res 2008, 36, 6418–6428. [Google Scholar] [CrossRef]
- Leger, A.J.; Mosquea, L.M.; Clayton, N.P.; Wu, I.H.; Weeden, T.; Nelson, C.A.; Phillips, L.; Roberts, E.; Piepenhagen, P.A.; Cheng, S.H.; et al. Systemic delivery of a Peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy. Nucleic Acid Ther 2013, 23, 109–117. [Google Scholar] [CrossRef]
- Gao, H. Progress and perspectives on targeting nanoparticles for brain drug delivery. Acta Pharm Sin B 2016, 6, 268–286. [Google Scholar] [CrossRef]
- Mae, M.; Langel, U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol 2006, 6, 509–514. [Google Scholar] [CrossRef]
- Faravelli, I.; Nizzardo, M.; Comi, G.P.; Corti, S. Spinal muscular atrophy--recent therapeutic advances for an old challenge. Nat Rev Neurol 2015, 11, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Feldkotter, M.; Schwarzer, V.; Wirth, R.; Wienker, T.F.; Wirth, B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 2002, 70, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Lorson, C.L.; Strasswimmer, J.; Yao, J.M.; Baleja, J.D.; Hahnen, E.; Wirth, B.; Le, T.; Burghes, A.H.; Androphy, E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 1998, 19, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Nakevska, Z.; Yokota, T. Challenges and future perspective of antisense therapy for spinal muscular atrophy: A review. Eur J Cell Biol 2023, 102, 151326. [Google Scholar] [CrossRef]
- Mercuri, E.; Darras, B.T.; Chiriboga, C.A.; Day, J.W.; Campbell, C.; Connolly, A.M.; Iannaccone, S.T.; Kirschner, J.; Kuntz, N.L.; Saito, K.; et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med 2018, 378, 625–635. [Google Scholar] [CrossRef]
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E.; et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med 2017, 377, 1723–1732. [Google Scholar] [CrossRef]
- Hammond, S.M.; Hazell, G.; Shabanpoor, F.; Saleh, A.F.; Bowerman, M.; Sleigh, J.N.; Meijboom, K.E.; Zhou, H.; Muntoni, F.; Talbot, K.; et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc Natl Acad Sci U S A 2016, 113, 10962–10967. [Google Scholar] [CrossRef]
- Shabanpoor, F.; Hammond, S.M.; Abendroth, F.; Hazell, G.; Wood, M.J.A.; Gait, M.J. Identification of a Peptide for Systemic Brain Delivery of a Morpholino Oligonucleotide in Mouse Models of Spinal Muscular Atrophy. Nucleic Acid Ther 2017, 27, 130–143. [Google Scholar] [CrossRef]
- Muntoni, F.; Wood, M.J. Targeting RNA to treat neuromuscular disease. Nat Rev Drug Discov 2011, 10, 621–637. [Google Scholar] [CrossRef]
- Henry, S.P.; Beattie, G.; Yeh, G.; Chappel, A.; Giclas, P.; Mortari, A.; Jagels, M.A.; Kornbrust, D.J.; Levin, A.A. Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int Immunopharmacol 2002, 2, 1657–1666. [Google Scholar] [CrossRef]
- Moulton, H.M.; Moulton, J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta 2010, 1798, 2296–2303. [Google Scholar] [CrossRef] [PubMed]
- Amantana, A.; Moulton, H.M.; Cate, M.L.; Reddy, M.T.; Whitehead, T.; Hassinger, J.N.; Youngblood, D.S.; Iversen, P.L. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjug Chem 2007, 18, 1325–1331. [Google Scholar] [CrossRef]
- Choi, J.M.; Ahn, M.H.; Chae, W.J.; Jung, Y.G.; Park, J.C.; Song, H.M.; Kim, Y.E.; Shin, J.A.; Park, C.S.; Park, J.W.; et al. Intranasal delivery of the cytoplasmic domain of CTLA-4 using a novel protein transduction domain prevents allergic inflammation. Nat Med 2006, 12, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Shin, J.H.; Sohn, M.H.; Harding, M.J.; Park, J.H.; Tobiasova, Z.; Kim, D.Y.; Maher, S.E.; Chae, W.J.; Park, S.H.; et al. Cell-permeable Foxp3 protein alleviates autoimmune disease associated with inflammatory bowel disease and allergic airway inflammation. Proc Natl Acad Sci U S A 2010, 107, 18575–18580. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, D.; Tae, H.S.; Gandotra, N.; Llopis, P.; Shen, N.; Altman, S. Basic peptide-morpholino oligomer conjugate that is very effective in killing bacteria by gene-specific and nonspecific modes. Proc Natl Acad Sci U S A 2011, 108, 16582–16587. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Clark, K.; Barton, C.; Tanguay, R.; Moulton, H. A Novel Zebrafish Model for Assessing In Vivo Delivery of Morpholino Oligomers. Methods Mol Biol 2018, 1828, 293–306. [Google Scholar] [CrossRef]
- Saar, K.; Lindgren, M.; Hansen, M.; Eiriksdottir, E.; Jiang, Y.; Rosenthal-Aizman, K.; Sassian, M.; Langel, U. Cell-penetrating peptides: a comparative membrane toxicity study. Anal Biochem 2005, 345, 55–65. [Google Scholar] [CrossRef]
- Sylvestre, M.; Lv, S.; Yang, L.F.; Luera, N.; Peeler, D.J.; Chen, B.M.; Roffler, S.R.; Pun, S.H. Replacement of L-amino acid peptides with D-amino acid peptides mitigates anti-PEG antibody generation against polymer-peptide conjugates in mice. J Control Release 2021, 331, 142–153. [Google Scholar] [CrossRef]
- Wu, R.P.; Youngblood, D.S.; Hassinger, J.N.; Lovejoy, C.E.; Nelson, M.H.; Iversen, P.L.; Moulton, H.M. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res 2007, 35, 5182–5191. [Google Scholar] [CrossRef]

| Drug Name (Market name) | ROA | Target gene | Indication | Modality | Chemistry | Mechanism of action | Approval | Company |
|---|---|---|---|---|---|---|---|---|
| Fomivirsen (Vitravene) | Intraocular | IE-2 mRNA | Cytomegalovirus (CMV) retinitis | ASO | 21mer PS DNA | RNase H1 | FDA/EMA (1998) | Ionis Pharmaceuticals, Novartis |
| Pegaptanib (Macugen) | Intraocular | Heparin-binding domain of VEGF-165 | Neovascular age-related macular degeneration | Aptamer | 27mer 2ʹ-F/2ʹ-OMe pegylated | Binding and blocking | FDA (2004) | OSI Pharmaceuticals |
| Mipomersen (Kynamro) | Subcutaneous | Apolipoprotein B100 | Homozygous familial hypercholesterolemia | ASO (gapmer) | 20mer PS 2ʹ-MOE | RNase H1 | FDA (2013) | Kastle Therapeutics, Ionis Pharmaceuticals, Genzyme |
| Eteplirsen (Exondys 51) | Intravenous | Exon 51 of DMD | Duchenne muscular dystrophy | ASO | 30mer PMO | Splicing modulation | FDA (2016) | Sarepta Therapeutics |
| Nusinersen (Spinraza) | Intrathecal | Exon 7 of SMN2 | Spinal muscular atrophy | ASO | 18mer PS 2ʹ-MOE | Splicing modulation | FDA/EMA (2016) | Ionis Pharmaceuticals, Biogen |
| Defibrotide (Defitelio) | Intravenous | Adenosine A1/A2 receptor |
Veno-occlusive disease in liver | Aptamer | Mixture of PO ssDNA and dsDNA | Binding and activating | FDA (2016) | Jazz Pharmaceuticals |
| Inotersen (Tegsedi) | Subcutaneous | Transthyretin | Polyneuropathy caused by hereditary transthyretin-mediated (hATTR) amyloidosis | ASO (gapmer) | 20mer PS 2ʹ-MOE | RNase H1 | FDA (2018) | Akcea Therapeutics |
| Milasen* | Intrathecal | CLN7 | Mila Makovec’s CLN7 gene associated with Batten disease | ASO | 22 mer 2′-O-MOE, PS, 5-methyl cytosine | Splicing modulation | FDA (2018) | Boston Children’s Hospital* |
| Patisiran (Onpattro) | Intravenous | Transthyretin | Polyneuropathy caused by hATTR amyloidosis | siRNA (LNP formulation) | 19 + 2mer 2ʹ-OMe modified | RNAi | FDA/EMA (2018) | Alnylam Pharma |
| Golodirsen (Vyondys 53) | Intravenous | Exon 53 of DMD | Duchenne muscular dystrophy | ASO | 25mer PMO | Splicing modulation | FDA (2019) | Sarepta Therapeutics |
| Givosiran (Givlaari) | Subcutaneous | 5-aminolevulinic acid synthase |
Acute hepatic porphyria (AHP) | siRNA (GalNAc conjugate) | 21/23mer Dicer substrate siRNA | RNAi | FDA/EMA (2019) | Alnylam Pharma |
| Volanesorsen (Waylivra) | Subcutaneous | Apolipoprotein C3 | Familial chylomicronemia syndrome (FCS) | ASO | 20 mer PS, 2ʹ-MOE | RNase H1 | EMA (2019) | Akcea Therapeutics |
| Viltolarsen (Viltepso) | Intravenous | Exon 53 of DMD | Duchenne muscular dystrophy | ASO | 21 mer PMO | Splicing modulation | FDA (2020) | NS Pharma |
| Casimersen (Amondys 45) | Intravenous | Exon 45 of DMD | Duchenne muscular dystrophy | ASO | 22 mer PMO | Splicing modulation | FDA (2021) | Sarepta Therapeutics |
| Tofersen (Qalsody) | Intrathecal | SOD1 | Amyotrophic lateral sclerosis | ASO | 20 mer 2ʹ-MOE, gapmer | RNase H1 | FDA (2023) | Ionis Pharmaceuticals, Biogen |
| Valeriasen | Intrathecal | KCNT1 | Epilepsy | ASO | 2ʹ-MOE, gapmer | RNase H1 | FDA (2020) | Boston Children’s Hospital* |
| Atipeksen | Intrathecal | ATM | Ataxia telangiectasia | ASO | Splicing modulation | Boston Children’s Hospital* |
| Bioconjugates | Brief introduction | Benefits |
|---|---|---|
| Lipid-based conjugates | Lipid-based moieties are usually cholesterol and its derivatives which are covalently conjugated to siRNA and antagomir ASOs to enhance the delivery. This group of bioconjugates enhances in vivo delivery by adhering to lipoprotein particles (such as HDL and LDL) in the circulation and therefore taking over the body’s natural system for lipid uptake and transport. The overall hydrophobicity of siRNAs governs their in vivo association with the various classes of lipoprotein, with the more hydrophobic conjugates preferentially attaching to LDL and primarily taken up by the liver. The less lipophilic conjugates preferentially bind to HDL and are consumed by the liver, adrenal glands, ovary, kidney, and small intestine. Another lipid derivatives α-tocopherol (vitamin E) was also found to increase the delivery of siRNA. |
|
| GalNac conjugates | Trimeric GalNac is the most clinically successful tissue-targeting ligand used in ASO delivery to date. GalNAc is a carbohydrate moiety that has a high affinity for the highly expressed asialoglycoprotein receptor 1 (ASGR1, ASPGR). This interaction promotes the endocytosis of PO ASOs and siRNAs into hepatocytes. Givosiran, a GalNAc-conjugated siRNA was granted FDA approval for the treatment of acute hepatic porphyria in November 2019 as a result its remarkable success. | |
| Antibody and Aptamer conjugates | Antibody–RNA bioconjugates offer a promising strategy for nucleic acid therapeutics, however, their utility for oligonucleotide delivery is still in the early stages of development. Antibodies are useful for the targeted delivery of oligonucleotides to cells or tissues that other methods cannot reach since they are very selective in recognizing target antigens. Similar to antibodies, aptamers bind to their respective target proteins with high affinity. Aptamers bind to their specific target proteins with high affinity, just like antibodies do. Aptamers are regarded as chemical antibodies and have demonstrated many advantages over antibodies, including being easier and less expensive to produce (i.e., through chemical synthesis), smaller size, and lower immunogenicity. | |
| Polymer conjugates | PEG is a non-ionic, hydrophilic polymer with a wide range of applications. It is widely used to prolong blood circulation time and improve drug efficacy. PEGylation, which involves covalently adding PEG to a drug, improves the stability of ASOs and reduces renal excretion by forming a protective hydration layer around them. PEG-conjugated drugs have been found to have better pharmacokinetic and pharmacodynamic properties in terms of the drug’s chemical aspects of absorption, distribution, metabolism, excretion, and toxicity (ADMET). Other polymers besides PEG have also received attention, including poly(glycerol), poly(2-oxazoline), poly (amino acid), and poly[N-(2-hydroxypropyl)methacrylamide] because they are more ADMET-enhancing and less immunogenic. | |
| Peptide-based conjugates | Peptides are short chains of amino acids that can serve as carriers for oligonucleotide delivery for its cell-specific targeting, cell-penetrating, or endosomolytic properties. More information about peptide conjugates is mentioned in section- 6.2.1. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




