Submitted:
24 July 2023
Posted:
25 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Indoor air sampling
2.2. Isolation and conservation of yeasts
2.3. Molecular identification, comparison of sequences and phylogenetic analyses
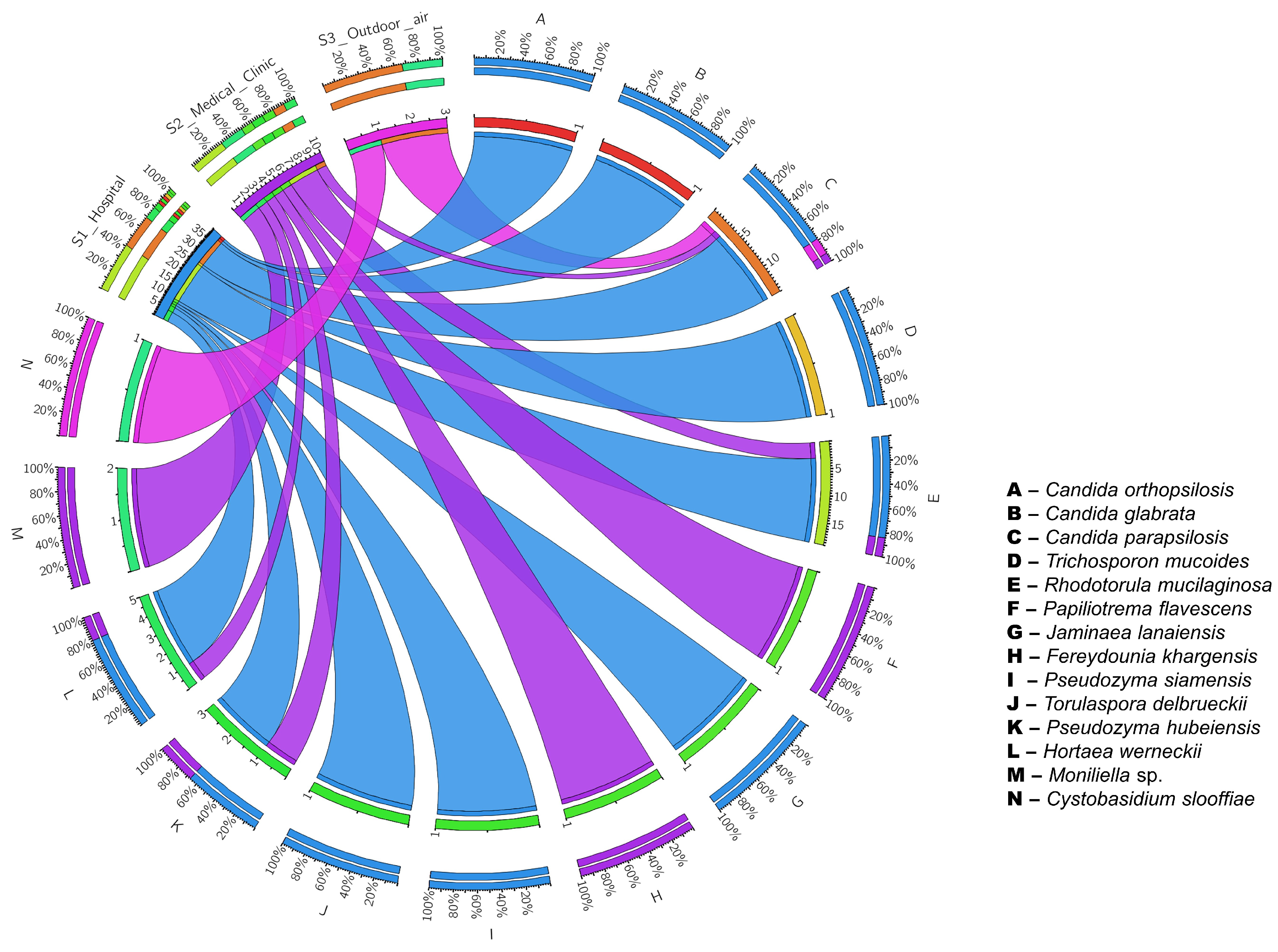
3. Results
4. Discussion
5. Conclusions
Author Contributions
Data Availability Statement
Conflicts of Interest
References
- Dadi, N.C.T.; Radochová, B.; Vargová, J.; Bujdáková, H. Impact of Healthcare-Associated Infections Connected to Medical Devices—An Update. Microorganisms. 2021, 9, 2332. [Google Scholar] [CrossRef]
- Klevens, R.M.; Edwards, J.R.; Richards, C.L.Jr.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef]
- Corrales-Fernández, M.J.; Gea-Velázquez de Castro, M.T.; Limón-Ramírez, R.; Miralles-Bueno, J.J.; Requena-Puche, J.; Aranaz-Andrés, J.M. Factores que contribuyen a la infección relacionada con la asistencia sanitaria: cómo evitarlos. Rev Calid Asis. 2011, 26, 367–375. [Google Scholar] [CrossRef]
- Suleyman, G.; Alangaden, G.J. Nosocomial Fungal Infections. Infect Dis Clin North Am. 2016, 30, 1023–1052. [Google Scholar] [CrossRef]
- Vazquez, J.A.; Miceli, M.H.; Alangaden, G. Invasive fungal infections in transplant recipients. Ther Adv Infect Dis. 2013, 1, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Presente, S.; Bonnal, C.; Normand, A.C.; Gaudonnet, Y.; Fekkar, A.; Timsit, J.F.; Kernéis, S. Hospital Clonal Outbreak of Fluconazole-Resistant Candida Parapsilosis Harboring the Y132F ERG11p Substitution in a French Intensive Care Unit. Antimicrob Agents Chemother. 2023, 16, e0113022. [Google Scholar] [CrossRef]
- Caggiano, G.; Lovero, G.; De Giglio, O.; Barbuti, G.; Montagna, O.; Laforgia, N.; Montagna, M.T. Candidemia in the Neonatal Intensive Care Unit: A Retrospective, Observational Survey and Analysis of Literature Data. Biomed Res Int. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lona-Reyes, J.C.; Gómez-Ruiz, L.M.; Cordero-Zamora, A.; Cortés-González, S.I.; Quiles-Corona, M.; Pérez-Ramírez, R.O.; Pinto-Macedo, H. Invasive candidiasis in a neonatal intensive care unit in Lagos, Nigeria. Niger Postgrad Med J. 2017, 24, 150–154. [Google Scholar]
- Ezenwa, B.N.; Oladele, R.O.; Akintan, P.E.; Fajolu, I.B.; Oshun, P.O.; Oduyebo, O.O.; Ezeaka, V.C. Invasive candidiasis in a neonatal intensive care unit in Lagos, Nigeria. Niger Postgrad Med J. 2017, 24, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.P.M.; Queijeiro-López, A.M.; Araújo, M.A.; Araujo, L.A.; Silva-Filho, E.A. Airborne Fungi in Indoor Hospital Environments. Int J Curr Microbiol App Sci. 2019, 5, 627–632. [Google Scholar] [CrossRef]
- Seervi, K.L.; Khatri, P.K.; Parihar, R.S.; Kulshrestha, S.; Meena, S.; Bora, A. Emergence of Drug Resistant Fungi in Hospital Air: A Need for Strict Microbiological Surveillance. Int J Curr Microbiol App Sci. 2016, 8, 2749–2772. [Google Scholar] [CrossRef]
- Mohajeri, P.; Soltani, S.; Getso, M.I.; Khatib, M.; Dastranj, M.; Farahani, A. Investigation of bio-air contamination in some hospitals of Kermanshah, Iran. Adv Hum Biol. 2019, 9, 65–70. [Google Scholar]
- valedeyni asl, F.; Arzanlo, M.; Fazlzadeh, M.; Amani, S.; Hazrati, S. Types and concentration of fungal bio-aerosols in hospital indoor air of Imam Khomeini and Alavi hospital in Ardabil city during 2016. IOH. 2017, 14, 103–113. [Google Scholar]
- Brown, C.S.; Guy, R. National Public Health Response to Candida auris in England. J Fungi. 2019, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Kenters, N.; Kiernan, M.; Chowdhary, A.; Denning, D.W.; Pemán, J.; Saris, K.; Schelenz, S.; Tartari, E.; Widmer, A.; Meis, J.F.; Voss, A. Control of Candida auris in healthcare institutions: Outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int J Antimicrob Agents. 2019, 54, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Agência Nacional de Vigilância Sanitária. Resolução-RE Nº 09. Ministério da Saúde do Brasil. 2003, 1–14.
- Silva-Filho, E.A.; Santos, S.K.B.; Resende, A.M.; Morais, J.O.F.; Morais, M.A.; Simões, D.A. Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting. Anton Leeuw Int J G. 2005, 88, 13–23. [Google Scholar] [CrossRef]
- Hsu, M.; Chen, K.; Lo, H.; Chen, Y.; Liao, M.; Lin, Y.; Li, S. Species identification of medically important fungi by use of real-time LightCycler PCR. J Med Microbiol. 2003, 52, 1071–1076. [Google Scholar] [CrossRef]
- Hesham, A.E.; Wambui, V.; Ogola, J.O.H.; Maina, J.M. Phylogenetic analysis of isolated biofuel yeasts based on 5.8S-ITS rDNA and D1/D2 26S rDNA sequences. J Genet Eng Biotechnol. 2014, 12, 37–43. [Google Scholar] [CrossRef]
- Staden, R.; Judge, D.P.; Bonfield, J.K. Analyzing Sequences Using the Staden Package and EMBOSS. In Introduction to Bioinformatics; Krawetz, S.A., Womble, D.D., Eds.; Humana Press: Totowa-NJ, United States, 2003; pp. 393–410. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J Mol Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Muhire, B.; Martin, D.P.; Brown, J.K.; Navas-Castillo, J.; Moriones, E.; Zerbini, F.M.; Rivera-Bustamante, R.; Malathi, V.G.; Briddon, R.W.; Varsani, A. A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol. 2013, 158, 1411–1424. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2018, 183, 591–596. [Google Scholar] [CrossRef]
- Veysi, R.; Heibati, B.; Jahangiri, M.; Kumar, P.; Latif, M.T.; Karimi, A. Indoor air quality-induced respiratory symptoms of a hospital staff in Iran. Environ. Monit. Assess. 2019, 191, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Qian, H.; Ye, J.; Sun, F.; Zhuge, Y.; Wang, S.; Liu, C.; Cao, G.; Zheng, X. Assessment of airborne bacteria and fungi in different-type buildings in Nanjing, a hot summer and cold winter moist Chinese city. Build Environ. 2021, 205, 108258. [Google Scholar] [CrossRef]
- Adams, R.I.; Bhangar, S.; Pasut, W.; Arens, E.A.; Taylor, J.W.; Lindow, S.E.; Nazaroff, W.W.; Bruns, T.D. Chamber bioaerosol study: outdoor air and human occupants as sources of indoor airborne microbes. PLoS One. 2015, 10, e0128022. [Google Scholar] [CrossRef] [PubMed]
- Zemouri, C.; Volgenant, C.M.C.; Buijs, M.J.; Crielaard, W.; Rosema, N.A.M.; Brandt, B.W.; Laheij, A.M.G.A.; De Soet, J. J. Dental aerosols: microbial composition and spatial distribution. J. Oral Microbiol. 2020, 12, 1762040. [Google Scholar] [CrossRef]
- Mirhoseini, S.H.; Koolivand, A.; Bayani, M.; Sarlak, H.; Moradzadeh, R.; Ghamari, F.; Sheykhan, A. Quantitative and qualitative assessment of microbial aerosols in different indoor environments of a dental school clinic. Aerobiologia (Bologna). 2021, 37, 1–8. [Google Scholar] [CrossRef]
- Venceslau, E.M.; Martins, R.P.P.; Oliveira, I.D. Frequency of airborne fungus in critical areas at hospital unit of Aracaju, Sergipe, Brazil. RBAC. 2012, 44, 26–30. [Google Scholar]
- Pedrosa, K.P.; do Nascimento, J.P.M.; Araújo, M.A.; Silva, M.S.S.; Santos, D.E.; Silva-Filho, E.A. Airborne Fungi in a Neonatal Intensive Care Unit and Operating Theater in a University Hospital. International Journal for Innovation Education and Research. 2022, 10, 41–55. [Google Scholar] [CrossRef]
- Yamin, D. 1.; Husin, A.; Harun, A. Distribution of candidemia in Malaysian tertiary care hospital revealed predominance of Candida parapsilosis. Trop. Biomed. 2020, 37, 903–910. [Google Scholar] [PubMed]
- Doi, A.M.; Pignatari, A.C.C.; Edmond, M.B.; Marra, A.R.; Camargo, L.F.A.; Siqueira, R.A.; Mota, V.P.; Colombo, A.L. Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian national surveillance program. PLoS One. 2016, 11, e0146909. [Google Scholar] [CrossRef]
- Souza, A.K.P.; Nascimento, J.P.M.; Araújo, M.A.S.; Pedrosa, K.P.S.; Tenorio, B.M.; Pires, L.L.S.; Lima, G.B.C.; Barboza, R.I.S.; Silva-Filho, E.A. Airborne Fungi in Neonatal Intensive Care Unit of a Public Hospital in Brazil. Int J Curr Microbiol App Sci. 2019, 8, 1210–1219. [Google Scholar] [CrossRef]
- Sudharsanam, S.; Swaminathan, S.; Ramalingam, A.; Thangavel, G.; Annamalai, R.; Steinberg, R.; Balakrishnan, K.; Srikanth, P. Characterization of indoor bioaerosols from a hospital ward in a tropical setting. Afr. Health Sci. 2012, 12, 217–225. [Google Scholar] [CrossRef]
- Diba, K.; Rhaimirad, M.; Makhdoomi, K.; Khorshidvand, Z.A. Identification of Candida species isolated from hospital acquired infections cases and hospital indoor environments. Afr. J. Microbiol. Res. 2012, 6, 4164–4168. [Google Scholar] [CrossRef]
- Kumar, M.R.; Prasad, S.R.; Urhekar, A.D. Incidence of Candida in Air from the Hospital Environment. Int J Adv Microbiol Health Res. 2017, 1, 29–33. [Google Scholar]
- Cordeiro, R.A.; Brilhante, R.S.N.; Pantoja, L.D.M.; Moreira-Filho, R.E.; Vieira, P.R.N.; Rocha, M.F.G.; Monteiro, A.J.; Sidrim, J.J.C. Isolation of pathogenic yeasts in the air from hospital environments in the city of Fortaleza, northeast Brazil. Braz J Infect Dis. 2010, 14, 30–34. [Google Scholar] [CrossRef]
- Sanna, C.; Marras, L.; Desogus, A.; Marras, B.; Montero, N.; Bertolino, G.; Schintu, M.; Coroneo, V. Evaluation of Rhodotorula spp. contamination in hospital environments. World J Pediatr Congenit Heart Surg. 2021, 193, 152. [Google Scholar] [CrossRef]
- Nogueira, N.; Júnior, E. Tinea Nigra na cidade de Campos dos Goytacazes, Rio de Janeiro. Revista Cienfica da FMC. 2012, 7, 20–24. [Google Scholar] [CrossRef]
- Tap, R.M.; Ramli, N.Y.; Sabaratnam, P.; Hashim, R.; Bakri, A.R.A.; Bee, L.B.; Ginsapu, S.J.; Ahmad, R.; Razak, M.F.A.; Ahmad, N. First Two Cases of Fungal Infections Associated with Multi-drug Resistant Yeast, Fereydounia khargensis. Mycopathologia. 2016, 181, 531–537. [Google Scholar] [CrossRef]
- Hunninghake, J.; Schuety, C.; Calvano, T.; Ferraro, D. Disseminated Trichosporon infection. Crit Care Med. 2019, 47, 270. [Google Scholar] [CrossRef]
- Rostami, N.; Alidadi, H.; Zarrinfar, H.; Salehi, P. Assessment of indoor and outdoor airborne fungi in an Educational, Research and Treatment Center. Ital. J. Med. 2017, 11, 52–56. [Google Scholar] [CrossRef]

| Species | Sequences (5’-3’)* | Size (pb) |
|---|---|---|
| Candida krusei | CKRU1:GCATCGATGAAGAACGCAGC CKRU2:AAAAGTCTAGTTCGCTCGGGCC | 258 |
| Candida albicans | CALB1:TTTATCAACTTGTCACACCAGA CALB2:ATCCCGCCTTACCACTACCG | 273 |
| Candida parapsilosis | CPA1:GCCAGAGATTAAACTCAACCAA CPA2:CCTATCCATTAGTTTATACTCCGC | 300 |
| Candida tropicalis | CTR1:CAATCCTACCGCCAGAGGTTAT CTR2:TGGCCACTAGCAAAATAAGCGT | 372 |
| Candida glabrata | CGL1:TTATCACACGACTCGACACT CGL2:CCCACATACTGATATGGCCTACAA | 423 |
| Sample | Environment/Collection site | Species |
|---|---|---|
| AYH01 | Hospital/Operating room | Candida parapsilosis |
| AYC02 | Medical Clinic/Doctor’s office | Candida parapsilosis |
| AYH03 | Medical Clinic/Doctor’s office | Candida parapsilosis |
| AYH04 | Hospital/Examination room | Candida parapsilosis |
| AYH05 | Hospital/Examination room | Candida parapsilosis |
| AYH06 | Hospital/Post-anesthesic care unit | Candida parapsilosis |
| AYC07 | Medical Clinic/Serology | Candida parapsilosis |
| AYC08 | Medical Clinic/Serology | Candida parapsilosis |
| AYC09 | Medical Clinic/Biochemistry laboratory | Candida parapsilosis |
| AYC10 | Medical Clinic/Collection room | Candida parapsilosis |
| AYH11 | Hospital/Urgency and Emergency | Candida parapsilosis |
| AYH12 | Hospital/Reception | Candida parapsilosis |
| AYH13 | Hospital/Reception | Candida parapsilosis |
| AYH14 | Hospital/Reception | Candida parapsilosis |
| AYH15 | Hospital/Waiting room | Candida parapsilosis |
| AYH16 | Hospital/Waiting room | Candida parapsilosis |
| AYH17 | Hospital/Waiting room | Candida parapsilosis |
| AYH18 | Hospital/Waiting room | Candida parapsilosis |
| AYH19 | Hospital/Ultrasound | Candida parapsilosis |
| AYH20 | Hospital/ICU | Candida parapsilosis |
| AYH21 | Hospital/Examination room | Candida parapsilosis |
| AYH22 | Hospital/Examination room | Candida parapsilosis |
| AYH23 | Hospital/Examination room | Candida parapsilosis |
| AYH24 | Hospital/Apartment | Candida parapsilosis |
| AYH25 | Hospital/Observation room | Candida parapsilosis |
| AYH26 | Hospital/Operating room reception | Candida parapsilosis |
| AYH27 | Hospital/Operating room reception | Candida parapsilosis |
| AYH28 | Hospital/Reception | Candida parapsilosis |
| AYH29 | Hospital/Diagnostic Center | Candida parapsilosis |
| AYH30 | Hospital/Diagnostic Center | Candida parapsilosis |
| AYH32 | Hospital/Operating room | Candida parapsilosis |
| AYH33 | Hospital/Operating room | Candida parapsilosis |
| AYH34 | Hospital/Hemodialysis | Candida parapsilosis |
| AYH35 | Hospital/Hemodialysis | Candida parapsilosis |
| AYH36 | Hospital/Hemodialysis | Candida parapsilosis |
| AYH37 | Hospital/Hemodialysis | Candida parapsilosis |
| AYH38 | Hospital/Chemotherapy | Candida parapsilosis |
| AYH39 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH40 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH41 | Hospital/Social assistance | Candida parapsilosis |
| AYH42 | Hospital/Application room | Candida parapsilosis |
| AYH43 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH44 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH45 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH46 | Hospital/Waiting room | Candida parapsilosis |
| AYH47 | Hospital/Clinical screening | Candida parapsilosis |
| AYH48 | Hospital/Occupational therapy | Candida parapsilosis |
| AYH49 | Hospital/Operating room | Candida parapsilosis |
| AYH50 | Hospital/Operating room | Candida parapsilosis |
| AYH51 | Hospital/Sterilization room | Candida parapsilosis |
| AYH52 | Hospital/Doctor’s office | Candida parapsilosis |
| AYH53 | Hospital/Application room | Candida parapsilosis |
| AYH54 | Medical Clinic/Quality control | Candida parapsilosis |
| AYH55 | Medical Clinic/Quality control | Candida parapsilosis |
| AYH56 | Medical Clinic/Recption | Candida parapsilosis |
| AYO01 | Outdoor air | Candida parapsilosis |
| AYO02 | Outdoor air | Candida parapsilosis |
| AYO03 | Outdoor air | Candida parapsilosis |
| AYO04 | Outdoor air | Candida parapsilosis |
| AYO05 | Outdoor air | Candida parapsilosis |
| AYO06 | Outdoor air | Candida parapsilosis |
| AYO07 | Outdoor air | Candida parapsilosis |
| Sample | Environment/Collection site | Species | Accession number |
|---|---|---|---|
| AYH57 | Hospital/Reception | Candida orthopsilosis | GenBank: MT001254 |
| AYC58 | Medical Clinic/Doctor’s office | Fereydounia khargensis | GenBank: MT001262 |
| AYC59 | Medical Clinic/Doctor’s office | Rhodotorula mucilaginosa | GenBank: MT001268 |
| AYH60 | Hospital/Operating room | Hortaea werneckii | GenBank: MT001237 |
| AYH61 | Hospital/Operating room | Hortaea werneckii | GenBank: MT001242 |
| AYC62 | Medical Clinic/Operating room | Rhodotorula mucilaginosa | GenBank: MT001269 |
| AYC63 | Medical Clinic/Doctor’s office | Candida parapsilosis | GenBank: MT001241 |
| AYC64 | Medical Clinic/Doctor’s office | Moniliella sp. | GenBank: MT001260 |
| AYC65 | Medical Clinic/Doctor’s office | Moniliella sp. | GenBank: MT001263 |
| AYH66 | Hospital/Procedures room | Rhodotorula mucilaginosa | GenBank: MT001270 |
| AYH67 | Hospital/Hemodialysis | Candida parapsilosis | GenBank: MT001243 |
| AYC68 | Medical Clinic/Reception | Pseudozyma hubeinsis | GenBank: MT001240 |
| AYC69 | Medical Clinic/Storage room | Papiliotrema flavescens | GenBank: MT001249 |
| AYC70 | Medical Clinic/Fractionation room | Hortaea werneckii | GenBank: MT001253 |
| AYC71 | Medical Clinic/Microbiology laboratory | Rhodotorula mucilaginosa | GenBank: MT001271 |
| AYH72 | Hospital/Observation room | Candida parapsilosis | GenBank: MT001244 |
| AYH73 | Hospital/Observation room | Hortaea werneckii | GenBank: MT001258 |
| AYH74 | Hospital/Observation room | Candida parapsilosis | GenBank: MT001245 |
| AYH75 | Hospital/Urgency and Emergency | Rhodotorula mucilaginosa | GenBank: MT001272 |
| AYH76 | Hospital/Neonatal ICU | Pseudozyma hubeiensis | GenBank: MT001247 |
| AYH77 | Hospital/Apartment | Rhodotorula mucilaginosa | GenBank: MT001273 |
| AYH78 | Hospital/Hemodialysis | Pseudozyma siamensis | GenBank: MT001251 |
| AYH79 | Hospital/Operating room | Candida glabrata | GenBank: MN966855 |
| AYH80 | Hospital/Post-anesthesia care unit | Candida parapsilosis | GenBank: MT001246 |
| AYH81 | Hospital/Storage room | Trichosporon mucoides | GenBank: MT001239 |
| AYH82 | Hospital/Audit | Torulaspora delbrueckii | GenBank: MT001238 |
| AYH83 | Hospital/Hemodialysis | Rhodotorula mucilaginosa | GenBank: MT001274 |
| AYH84 | Hospital/Observation room | Rhodotorula mucilaginosa | GenBank: MT001275 |
| AYH85 | Hospital/Waiting room | Rhodotorula mucilaginosa | GenBank: MT001276 |
| AYH86 | Hospital/Nurses station | Rhodotorula mucilaginosa | GenBank: MT001277 |
| AYH87 | Hospital/Pharmacy | Candida parapsilosis | GenBank: MT001248 |
| AYH88 | Hospital/Examination room | Candida parapsilosis | GenBank: MT001250 |
| AYH89 | Hospital/Social assistance | Candida parapsilosis | GenBank: MT001252 |
| AYH90 | Hospital/Macroscopy | Rhodotorula mucilaginosa | GenBank: MT001278 |
| AYH91 | Hospital/Macroscopy | Rhodotorula mucilaginosa | GenBank: MT001279 |
| AYH92 | Hospital/Clinical screening | Rhodotorula mucilaginosa | GenBank: MT001280 |
| AYH93 | Hospital/Occupational therapy | Rhodotorula mucilaginosa | GenBank: MT001281 |
| AYH94 | Hospital/Observation room | Candida parapsilosis | GenBank: MT001255 |
| AYH95 | Hospital/Doctor’s office | Candida parapsilosis | GenBank: MT001256 |
| AYH96 | Hospital/Administrative | Rhodotorula mucilaginosa | GenBank: MT001282 |
| AYH97 | Hospital/Application room | Pseudozyma hubeiensis | GenBank: MT001267 |
| AYH98 | Hospital/Application room | Candida parapsilosis | GenBank: MT001257 |
| AYH99 | Hospital/Telephone exchange | Candida parapsilosis | GenBank: MT001261 |
| AYH100 | Hospital/Coordination | Hortaea werneckii | GenBank: MT001259 |
| AYH101 | Hospital/Information technology | Jaminaea lanaiensis | GenBank: MT001264 |
| AYH102 | Hospital/Observation room | Rhodotorula mucilaginosa | GenBank: MT001283 |
| AYO08 | Outdoor air | Cystobasidium slooffiae | GenBank:MT001284 |
| AYO09 | Outdoor air | Candida parapsilosis | GenBank:MT001265 |
| AYO10 | Outdoor air | Candida parapsilosis | GenBank:MT001266 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





