Introduction
From the beginning of the coronavirus disease 2019 (COVID-19) caused by the pandemic outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Wuhan, China late 2019, reports on the severe gaps in vaccine and therapeutics have come in from all over the world. With about 652 million confirmed cases and more than six million documented deaths, the disease’s worldwide mortality and morbidity are still rising [
1]. The evolutionary dynamics of SARS-CoV-2 epidemics vary worldwide and depend on a range of variables, including host genetics, virus genetics, co-infection, environmental factors, country health infrastructures, and public attitudes and preventative strategies [
2,
3,
4,
5,
6]. However, drug discovery and immunization programs at the international level continue to take precedence to control and prevent the disease. The SARS-CoV-2, is a large, positive-sense, single-stranded, RNA coronavirus. The virus produces four major structural proteins: spike (S), nucleocapsid (N), envelope (E), and membrane (M). The spike protein mediates viral infection of cells via the transmembrane zinc protein angiotensin-converting enzyme 2 (ACE2) receptor. Structurally, the S protein is a type-I membrane trimer protein constructed from a homodimer of two subunits (S1 and S2) linked by a third membrane-embedded serine 2 protease subunit [
7]. The receptor-binding domain (RBD) is located at the top of the S1 subunit. The spike protein is highly glycosylated (O- and N-glycosylation); each monomer contains 22 N-glycosylation points, partially facilitating viral virulence. Furthermore, the N-glycans at the protein’s surface can mediate RBD protection against neutralization antibodies [
8]. The N-glycan structures are synthesized from glycan core Glc3Man9GlcNAc2 precursor, which is then processed into three types; oligo-mannose (2HexNAc), hybrid (3HexNAc), and complex (< 3HexNAc) structures [
9]. Many drugs have been tested for their ability to inhibit the spike protein-angiotensin converting enzyme-2 (ACE-2) interactions; they exert their potency by either blocking the RBD of the S protein [
10] or obstructing the ACE2 binding domain [
11]. Thus, despite enormous efforts being made for suitable candidates, there is yet significant gap closure remains in this area. While many products were developed, they were limited by global availability and/or other pitfalls in design.
Lectins are carbohydrate-binding proteins with high specificity and avidity that bind carbohydrate moieties reversibly without changing their chemical structures [
12]. Over the years, plant lectins have been extensively studied for their application as antimicrobial, anticancer, anti-inflammatory, antinociceptive agents, etc. [
13,
14,
15]. Several plant lectins have been identified as SARS-CoV-2 inhibitors. They are thought to bind the N-glycan structure near the RBD, thereby preventing the S protein from binding the ACE2. These interactions may also result in conformational adjustments that facilitate antibodies’ access to the target’s concealed epitopes and lead to immunological neutralization [
14,
16,
17]. Man-specific lectins belonging to the families’ legume (i.e., ConA Canavalia ensiformis and LcA Lens culinaris), Jacalin (i.e., BanLec Musa acuminata), Nictaba (i.e., Nictaba Nicotiana tabacum), Ricin- B (i.e., IraB Iris hollandica), and GNA (i.e., Gastrodianin Gastrodia elata) were reported as coronaviruses’ spike protein blockers that interact with the oligo-mannose structures located near the RBD [
18]. Instead of the typical carbohydrate-protein interactions, lectins can also engage in protein-protein interactions with the spike protein. A chitin-specific lectin Urtica dioica Agglutinin (UDA) isolated from the rhizomes of the Urtica dioica inhibits SARS-CoV-2 infection by binding to the RBD of the spike protein in a protein-protein manner [
19].
In this in-silico study, we investigated members of the Hevein-like lectins (chitin-specific lectins) identified from the genome of the spike moss plant (Selaginella moellendorffii, Family: Selaginellaceae) to study their interactions with the spike protein’s N-glycans and block the RBD, hence acting as potential SARS-CoV-2 inhibitors. To acquire a better understanding of the physical underpinnings of the complexes, the modelled proteins were docked against the RBD of the spike protein. The complexes were then further examined using molecular dynamic simulation on the iMODS server to provide an overview on the physical bases of the complexes. Because of its bioactive metabolites (primary and secondary), which are employed in both conventional and modern medicine as antifungal, antibacterial, antiviral, anticancer, anti-inflammatory, anti-allergy, and antioxidant agents, the spike moss was selected as a prospective source for lectin [
20]. A comprehensive overview of the domain architecture, gene architectures, genomic expansion, evolutionary relationship, and expressional profiles in various tissues and organs will also be provided by the study.
2. Materials and Methods
2.1. Screening the spike moss genome for putative Hevein-like lectin genes
The genome assembly (scaffold level) of Selaginella moellendorffii (v1.0, ID:88036) [
21] found in Phytozome v13 the plant genomics resources, [available at (
https://phytozome-next.jgi.doe.gov/) accessed April 15, 2023] [
22] was searched for Hevein-like lectin genes by aligning the proteome against the sequence of Hevea brasiliensis agglutinin (Hevein – GenBank: ABW34946.1, Pfam: PF00187) using Phytozome-BLASTP tool (E value <0.0001, Matrix: BLOSUM62). The sequence with the highest identity was used for a second BLAST search. The candidate Hevein-like lectin sequences obtained was then retrieved using the Phytozome-BioMart tool, and each sequence was checked for the presence of at least one Hevein-like domain using the InterProScan 5 [
23] [available at (
http://www.ebi.ac.uk/interpro/) accessed April 15, 2023]. The spike moss Hevein-like putative genes structure was studied by investigating the exon/intron organization of each gene coding sequence (CDS) in reference to their genomic DNA sequences using the Gene Structure Display Server (GSDS) [available at (
http://gsds.gao-lab.org/) accessed April 15, 2023] [
24] (Guo et al., 2007).
2.2. Analysis of Hevein-like gene expansion and evolutionary relationship
The Hevein-like putative lectin gene IDs were checked against the Pant Duplicate Gene Database PlantDGD [available at (
http://pdgd.njau.edu.cn:8080/)accessed April 15, 2023] [
25]to identify the type of duplication and the duplicate genes, then the synonymous substitution (Ks) was calculated using the Ka/Ks calculator found in the TBtools v1.0986853 [
26], values higher than 1 were excluded. The phylogenetic trees were constructed using aligned trimmed lectin-like domain sequences. The maximum likelihood method and the JTT matrix-based substitution model were used in MEGA X to analyze the evolutionary relationship [
27]. The final bootstrap for consensus trees was inferred from 1000 replicates.
2.3. Expressional profile of Hevein-like genes based on publicly available resources
The dataset of expression profiling of Selaginella moellendorffii analyzed by high throughput sequencing and deposited in the NCBI Gene Expression Omnibus (GEO) (GSE123120) was downloaded [
28]. The GSE123120 file containing RNA-seq expression raw read counts per gene for semplice replicates (seeds, rhizophore, leaf, and shoot) was selected for Hevein-like genes read counts. Data was normalized and the differential expression was analyzed by the Integrated Differential Expression and Pathway analysis [available at (
http://bioinformatics.sdstate.edu/idep96/) accessed April 15, 2023] (iDEP, v0.96 [
29]. Each Hevein-like coding sequence (CDS) was searched for miRNA targeting sites using the Plant Small RNA Target Analysis Server (psRNATarget, v2) under default setting [available at (
https://www.zhaolab.org/psRNATarget/analysis?function=1)accessed April 15, 2023] [
30].
2.4. Characterization of Hevein-like homologs
The protein sequence of Hevein-like lectin homologs were checked for the presence of signal peptide and transmembrane domain sequences using the SignalP 5.0 server [
31] [available at (
https://services.healthtech.dtu.dk/service.php?SignalP-5.0)accessed April 15, 2023] and TMHMM 2.0 [
32] available at (
https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) accessed April 15, 2023], respectively. Furthermore, each sequence was analyzed for the presence of potential glycosylation sites for N- glycan using NetNGlyc 1.0 server [
33] (available at
https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0) accessed April 15, 2023] and O-glycan using NetOGlyc 4.0 server [available at (
https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0) accessed April 15,2023] [
34]. The subcellular localization was predicted using the WoLF PSORT webserver [
35] [available at (
https://wolfpsort.hgc.jp/), accessed April 15, 2023], followed by the prediction of signal classes (signal peptide (SP), transmembrane (TM), intracellular (IC), and unconventional secretion (USP)) using the OutCyte 1.0 server available at (
http://www.outcyte.com/) accessed April 15, 2023 [
36].
2.5. Secondary structure prediction, structural modelling, and validation of Hevein-like homologs
The secondary structure of the 10 Hevein-like sequences were determined using the SOPMA available at (
https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/ npsa_sopma.html). accessed April 15, 2023] [
37]. The 3D protein structures of the lectin sequences were built using the transform-restrained Rosetta (trRosetta) webserver [available at (
https://yanglab.nankai.edu.cn/trRosetta/) accessed April 15, 2023] [
38]. The structures obtained by trRosetta were validated by examining the Phi/Psi Ramachandran plot using SAVES v6.0 Structure Validation Server [available at (
https://saves.mbi.ucla.edu/) accessed April 15, 2023 [
39] acquired from the PROCHECK server [
40], which evaluates the stereo-chemical properties of the modelled protein structures of lectins. Additionally, the PROSA (Protein Structure Analysis) webserver [available at (https: //prosa.services. came.sbg.ac.at/prosa.php) accessed April 15, 2023] [
41] was employed to examine the energy specifications of the protein models compared to database structures.
2.6. Retrieval and pre-processing of SARS-CoV-2 spike glycoprotein, and the lectins structures
The 3-dimensional (3D) crystal structure of the SARS-CoV-2 S glycoprotein, at a 2.80 Å resolution (total structure weight 438.26 KDa), was downloaded from the PDB databank [
42] with PDB ID: 6VXX [
43]. The Schrödinger suite’s Protein Preparation Wizard tool of Maestro software (Schrödinger Release 2021-2: Maestro) was used to pre-process and prepare the structure of the SARS-CoV-2 spike glycoprotein (PDB: 6VXX, designated as chain B) and the 10 Hevein-like structures. In this process, bonds order was assigned, hydrogens were added, missing loops and side chains were fixed, and the uncapped N-termini and C-termini were capped with ACE (N-acetyl) and NMA (N-methyl amide) groups, respectively. The sugar cofactor, N-acetylglucosamine (NAG) was retained in the protein structure. Afterwards, the protein was optimized using PROPKA at a pH of 7.4 and energy was minimized using the OPLS4 force field [
44].
2.7. Identifying the binding sites of the Spike glycoprotein and lectins for macromolecular and ligand docking
2.8. In silico molecular docking
The HDOCK server [available at (
http://hdock.phys.hust.edu.cn/)accessed April 15 2023, [
46] was used for in silico macromolecular docking of the lectins to N-linked glycans of SARS-CoV-2 spike glycoprotein using a hybrid algorithm approach. The HDOCK server uses an FFT-based method to perform global docking using the input receptor and ligand structure to sample acceptable binding conformations. Furthermore, the HDOCK docking tool performs rigid body docking by considering both the protein and the ligand (protein) onto grids. The binding energy of macromolecules was used to evaluate their binding mode and rank them based on their docking energies. In the selection process, lectin-RBD complexes were selected based on two factors: the docking lowest energy and, second, the interaction with N-acetylglucosamine (NAG) that contributed to the best complex selection when using the PDBsum: Structural summaries of PDB entries web server [available at (
http://www.ebi.ac.uk/pdbsum)accessed April 15,2023} [
40]. The HawkDock server (
http://cadd.zju.edu.cn/hawkdock/) [
47] was used to calculate the MM/GBSA (Molecular Mechanics-Generalized Born Surface Area) free binding energy for the docked complexes of lectins with SARS-CoV-2 S glycoprotein. The MM/GBSA was calculated based on the ff02 force field, the implicit solvent model, and the GBOBC1 model (interior dielectric constant = 1). The whole system was minimized for 5000 steps with a cut-off distance of 12 Å for van der Waals interactions (2000 cycles of steepest descent and 3000 cycles of conjugate gradient minimizations) and expressed in kcal mol-1. The docking modes generated were analysed and visualized using Discovery Studio 2021 [
48] which were subsequently exported as an image.
2.9. In silico mutant spike protein interactions
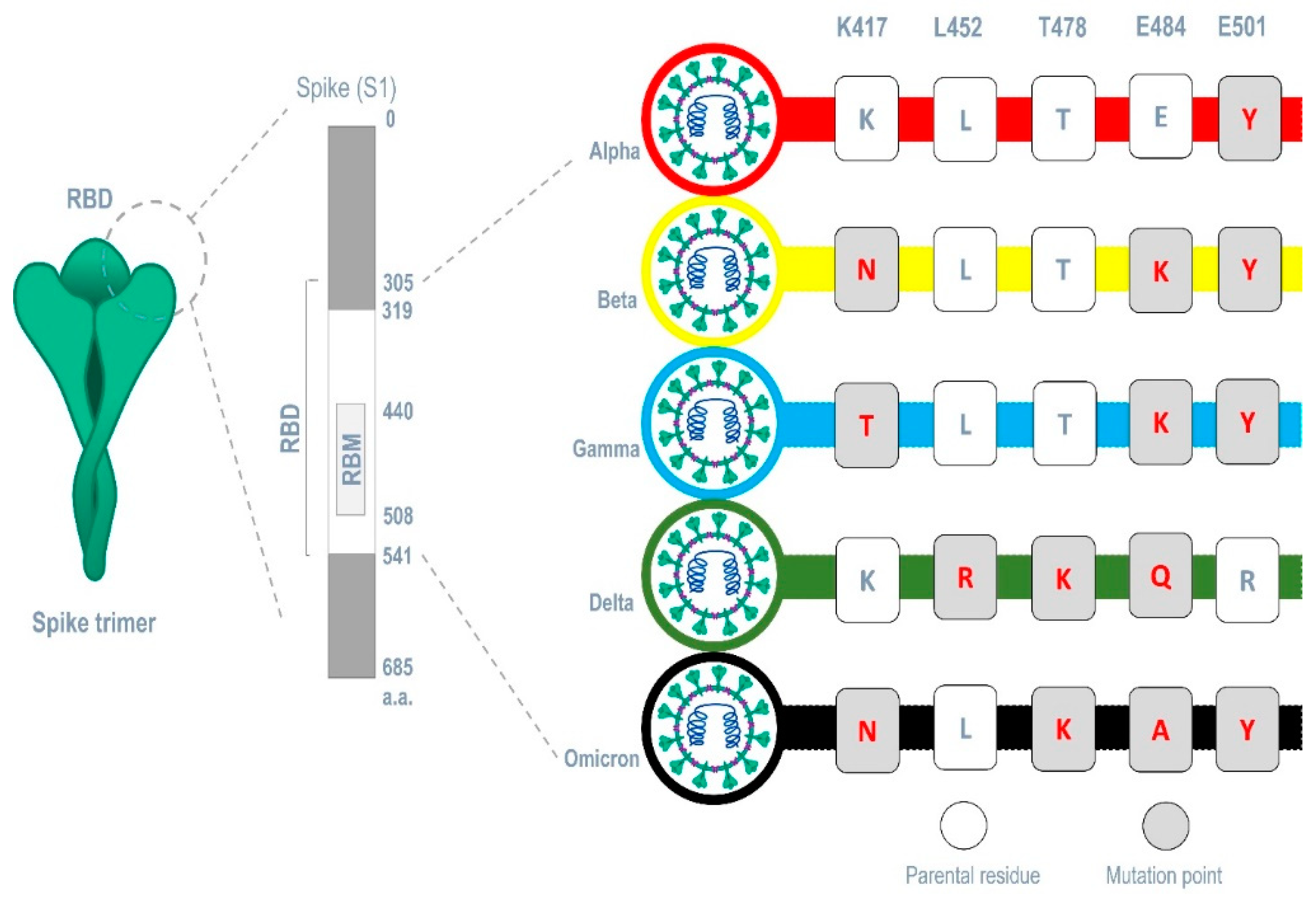
Five mutation points (i.e., Lys417Asn, Leu452Arg, Thr478Lys, Glu484Lys, and Asn501Tyr) are reported in spike protein’s ACE 2-receptor binding motif (RBM) from many SARS-CoV-2 variants were affected in the parental structure (PDB 6VXX) separately and five mutant structures were obtained, and each was docked against Smo446851, Smo125663, and Smo99732. The MM/GBSA free binding energy for each complex was calculated. Different combination of these mutations also occur within the RBM from the variants (alpha, beta, gamma, delta, and omicron) were also used to modify the spike protein parental structure and were docked with the aforementioned lectins.
2.10. Hot-spot analysis of the SARS-COV-2 RBD-Lectin complex
The energetically significant hotspot residues at the interface region of SARS-CoV-2 S glycoprotein and lectins were identified using the KFC Server (Knowledge-based FADE and Contacts) [available at (
https://mitchell-web.ornl.gov/KFC_Server/index.php), accessed on April 15, 2023] [
49].
2.11. Normal-State Analyses via Torsional Coordinate-Association
The iMODS online server [available at (
http://www.imods.chaconlab.org/), accessed on April 15, 2023] [
50] was used to investigate the collective flexibility/motion functions of the lectin–Spike glycoprotein complex based on normal-state analyses of their respective internal dihedral angle (torsional) coordinates [
51]. Within the PDB file of lectin–Spike glycoprotein docked complexes; the atoms/residues were continuously indexed. The complexes were uploaded to iMODS online server, parameters were kept as default, and the collective motions of proteins were investigated using normal mode analysis.
3. Results
3.1. General overview of Hevein-like lectins
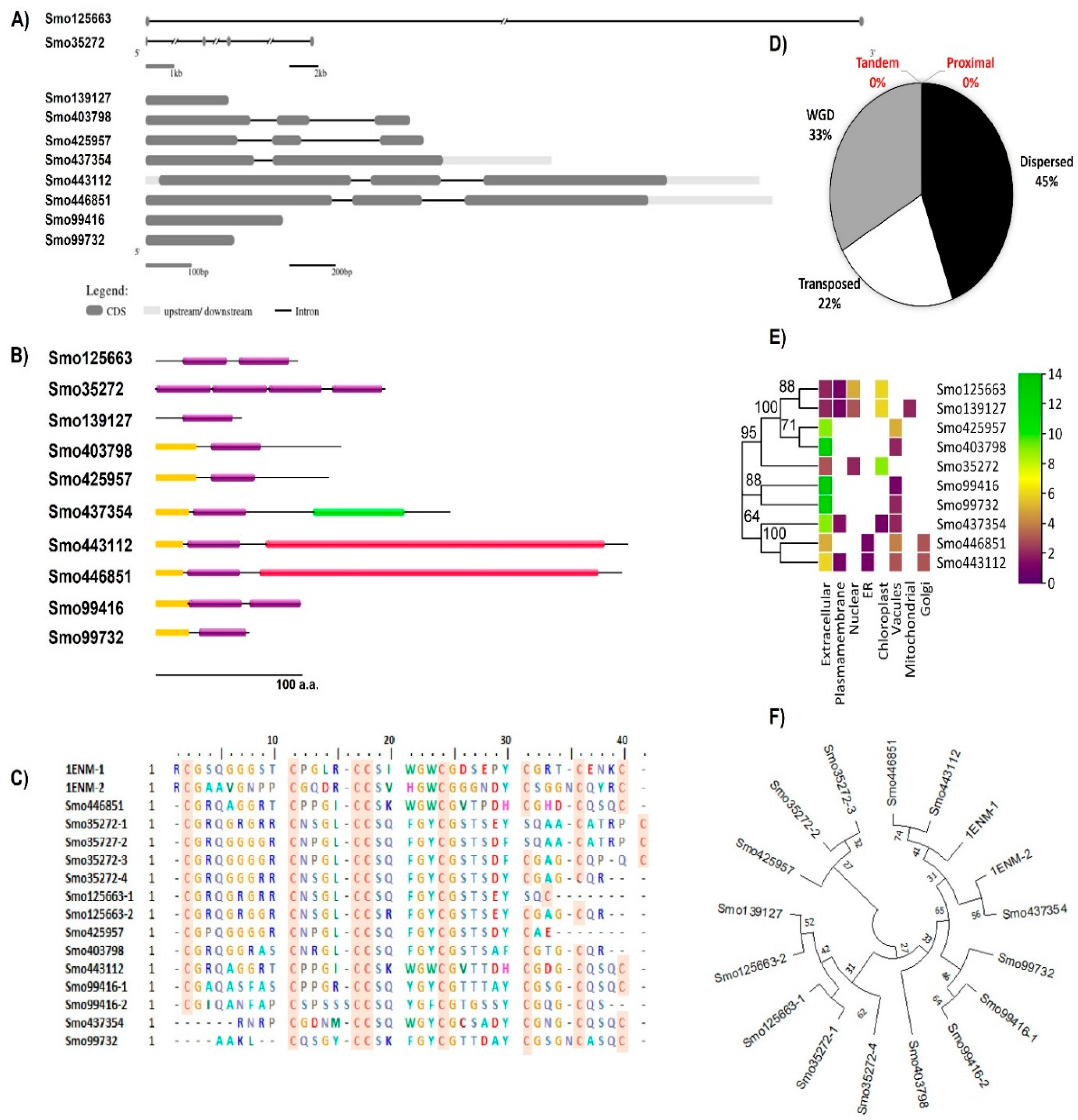
Searching the Selaginella moellendorffii (v1.0, ID:88036) genome for Hevein-like homologs resulted in identifying 10 sequences located in different scaffolds. A 30% of the identified lectins were secreted O-glycosylated chimerolectins. In these the Hevein domain fused by the C-terminal to a chitinase class I domain (PF00182) (Smo443112 and Smo446851, ~ 34 KDa) translated from three exons in each gene or to a lytic transglycosylase domain (PF03330) as in the 20.9 KDa sequence Smo437354. Merolectins (40%) and Hololectins (30%) were also reported. For instance, the Smo425957 duplicated by a transposition mechanism from the Smo403798 were both Merolectins predicted as extracellular proteins. These were attached by the N-terminal to a signal peptide and by the C-terminal to a stretch of the non-conserved region of <50 amino acids in length. Their respective genes were constructed from three exons interrupted by two introns. Two and four tandemly arrayed Hevein-like domains shared an evolutionary relationship with other Merolectins namely Smo139127, Smo425957, and Smo99732. Domains from Smo35272 4-domain Hololectin were believed to be expanded from Smo139127 by the mechanism of dispersion and from Smo125663 by the mechanism of wide genome duplication (WGD). One of the main characteristics of the Hevein domains is the presence of the eight highly conserved cysteine residues. At least one domain in the Hololectins contains all eight residues (
Table 1,
Figure 1).
3.2. Expression profile of Hevein-like genes in different organs
The transcription profile of Hevein-like members of the S. moellendorffii varies among members within and between the same tissue or organs analyzed. For instance, the expression level of Smo125663 in seeds, leaves, and shoot is ~ 4 folds higher than in rhizophore. However, Smo99416, Smo99732, and Smo139127 expression levels are generally lower in all tissues or organs compared to the rest of the other Hevein-like genes. Moreover, the Smo99416 transcript is the only one that has a targeting cleavage site for miRNA silencing and it is targeted by a single miRNA from the family MIR1082 (smo-niR1082a, located in scaffold 19) ( see Supplementary file 1
Figure S1 for more details).
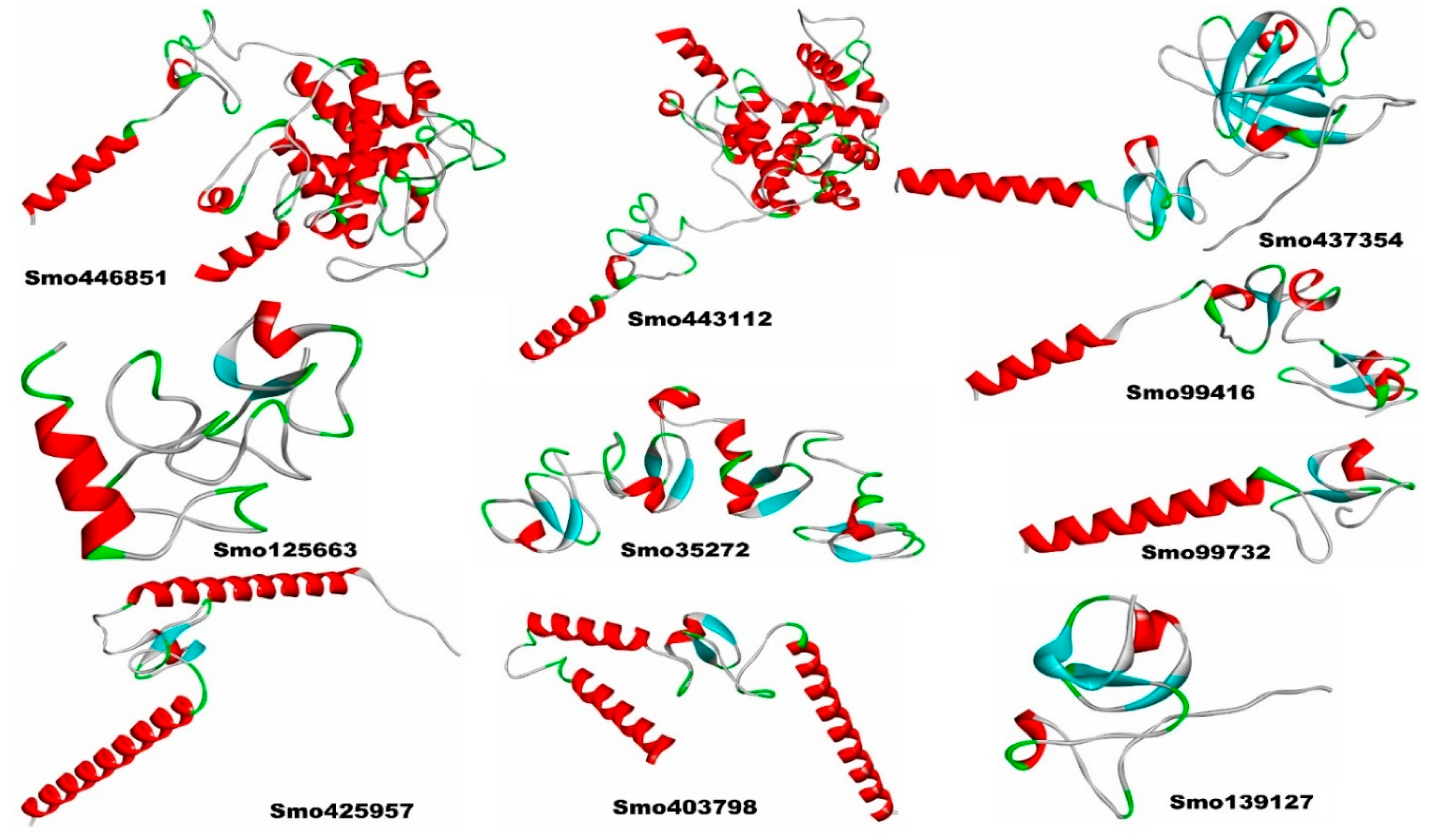
3.3. Structural model building of the lectins and their secondary structures.
The 3D structural models of Smo446851 and Smo443112 lectin sequences were built with the restraints from both deep learning and homologous templates of known X-ray structures of the class-1 chitinase (Glyco_hydro_19) lectin; 2DKV [
52], 3W3E [
53], 4TX7 [
54], 6LNR [
55], and 1DXJ [
56] using trRosetta webserver. Moreover, trRosetta proposed other templets for Smo437354 having confidence scores > 99, but rather lower identity scores in relation to the templates Expansin-like proteins (PDB ID: 3D30) [
57], (Beta-expansin 1a (EXPB1) (PDB ID: 2HCZ_X) [
58], Pollen allergen Phl p 1 (PDB ID: 1N10) (Fedorov 2003) [
59], and cellulose binding proteins (PDB ID: 4JS7 and 4JJO) (Yennawar 2013) [
60]. All templates were detected by running HHsearch against the PDB70 database, and confidence, coverage, identity, E-value, and Z-score scores were reported (
Figure 2, also see Supplementary file 1
Table S1). The other seven lectin model structures were constructed based on de novo folding guided by deep learning restraints using the trRosetta webserver. The TM scores for Smo446851 and Smo443112 were 0.88 and 0.86, respectively; whereas, the slightly lower scores ranging from 0.86 to 0.44 were predicted for the other lectins. The TM-score is a measure of confidence estimation that falls between 0 and 1 where high scores indicate a correctly predicted topology [
38] (this is detailed in Supplementary file 1
Table S2). The Ramachandran plots showed that >80% of the amino acid residues of Smo446851, Smo35272, Smo425957, Smo403798, Smo443112, Smo99416, Smo437354, Smo99732, fell within the allowed region; whereas, only 73.6% and 77.3% of the amino acids from the Smo125663 and Smo139127 were located within the allowed region. Furthermore, a very small percentage of the amino acids were located with the disallowed region, i.e., Smo446851 (0.8%), Smo125663 (2.8%), Smo403798 (0.9%), Smo443112 (0.8%), and Smo437354 (0.6%). However, these residues were not located within the Hevein domain and thus were not involved in the carbohydrate interaction (Supplementary file 1 Figures S2, S3-A and S3-B).
The analysis of the Hevein-like homolog sequences using SOPMA demonstrated the presence of α-helices (Hh), β-turns (Tt), random coils (Cc), and extended strand (Ec) (Supplementary file 1
Figure S4). Almost all of the lectins’ secondary structures exhibited a maximum frequency of random coils (Cc), except for Smo425957 and Smo403798, indicating protein high stability and flexibility [
61]. Moreover, Smo425957 and Smo403798 with a higher proportion of α-helices, accounted for 54.24 % and 39.52%, respectively. Alpha helices were more likely to be found in most thermophilic proteins [
62].
3.4. Identifying the binding sites of the S glycoprotein and lectins for macromolecular and ligand docking
In the receptor binding domain (RBD) of the S glycoprotein (PDB ID: 6VXX), the amino acid residue Lys436, Gly465, Tyr468, Tyr472, Leu474, Phe475, Ala494, Phe505, Asn506, Tyr508, Gln512, Gly515, Gln517, Thr519, Asn520, Gly521, Tyr524, and the cofactor N-glycan NAG1321 (located at N165) were identified as the binding site for macromolecular (protein-protein) docking with lectins and ligand docking of cofactor NAG with the lectins, respectively. Details of the binding site residues of all the lectins considered for macromolecular docking were summarized in Supplementary file 1
Table S3.
3.5. Molecular docking of lectins with SARS-CoV-2 spike protein
Since it has been previously established that spike protein monomer has 22 N-glycosylation 2 O-linked glycan points [
15,
63,
64] and that Hevein-like lectins are specific for N-acetylglucosamine and chitin (polymer of GlcNAc) [
65] 11 points of hybrid and complex N-glycans were of interest for the docking in this study. Due to their proximity to the ACE-2 RBD, the residues Asn165, Asn331, and Asn343 connected to complex N-glycan were of particular interest. The ACE-2 binding pocket and its surface N-glycans were studied for protein-protein and carbohydrate-protein interactions against each lectin, respectively.
We report that only three Hevein-like lectins (Smo35272, Smo425957, and Smo403798) interacted directly with the RBD residues in a protein-protein manner with binding free energy equal to -50.07, -16.12, and -22.72 Kcal/mol, respectively. However, neither of them interacted with the key residues of the receptor binding motif of the ACE2 (i.e., active key residues). Both Smo35272 and Smo403798 formed salt bridges with spike protein outside the vicinity of the RBD (Smo35272; Arg9 ⸺ Glu196 (3.2 Å), Arg47 ⸺ Glu132 (3.3 Å), and Arg154 ⸺ Glu298 (3.5 Å)) and (Smo403798; Arg72 ⸺ Asp198 (3.2 Å)). Smo35272 formed the highest number of hydrogen bonds (20 bonds) with the spike protein, 40% of them were established within the proximity of the RBD (residue span 320 – 385).
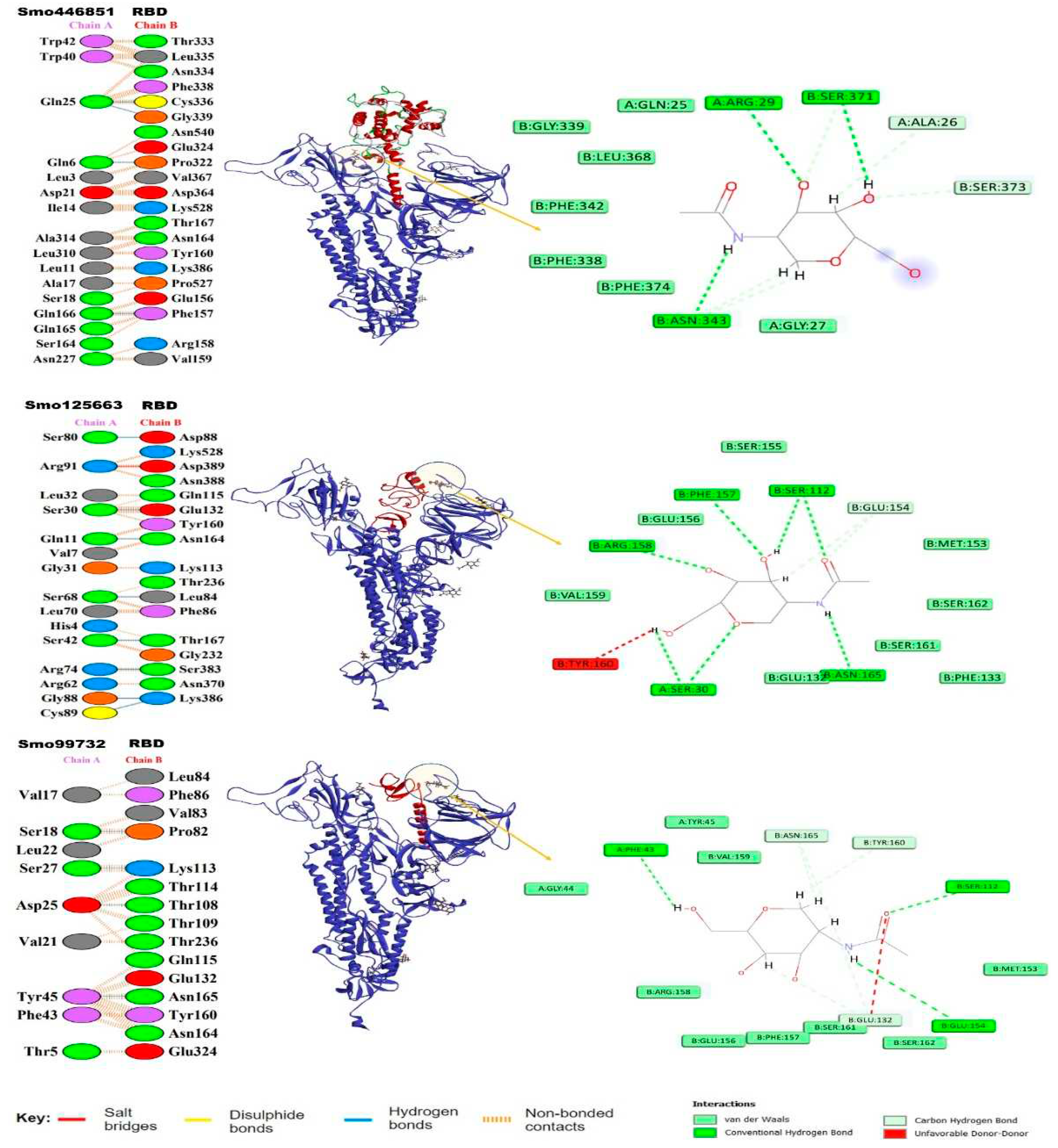
Our in silico molecular docking showed that only three lectins (Smo446851, Smo125663, and Smo99732) were able to interact with the NAG cofactors of the spike protein S1 via hydrogen bonding. In this, two hydrogen bonds were formed between Smo446851 and NAG1307 (located at Asn343); whereas, and a single hydrogen bond was observed between lectins Smo125663, and Smo99732 and the N-glycan NAG1321 (located at Asn165), respectively (
Table 2). These Hevein-like lectins which interact with the spike protein in a carbohydrate-protein manner have also several other amino acids that interact directly with the residues located in the vicinity of the RBD. Among the 16 interacting residues of Smo446851, only Gln25 of the Hevein domain formed two hydrogen bonds with the residues from the spike protein: Cys336 and Gly339. Based on the PPI generated by the PDBSum server, 12 of the 15 interacting residues from Smo125663 located in the Hevein-domains (region 19 – 49 and 57 – 91) showed good interaction with the RBD residues, of which three hydrogen bonds were formed with two residues from RBD (Ser383 and Lys386). Moreover, a salt bridge interaction was also reported between Arg91 from Smo125663 and ASP389 from the RBD. A lower number of interacting residues were observed for Smo99732 (9 amino acids), and only two residues of which Phe43 and Tyr45 are in the Hevein domain. Tyr45 shared a hydrogen bond with key residue Asn165 from the RBD (a glycosylation point for complex N-glycan) (
Figure 3). Furthermore, the estimated Generalized Born Model and solvent accessibility (MM/GBSA) calculations of lectins with S glycoprotein of docked complexes provided predictions for the binding energy and detailed the contributions to binding free energy per residue to assist in the analysis of binding structures in proteins by considering the electrostatic potentials (ELE), the Van der Waals potentials (VDW), the polar solvation (GB), and the nonpolar contribution to the solvation (SA) [
47]. The Smo99732 complexed with S protein has a lower MM/GBSA score (-26.5 Kcal/mol) compared to the complexes with Smo446851 (-17.5 Kcal/mol) and Smo125663 (-13.0 Kcal/mol). No favorable interactions were detected between the interactive key residues at the RBM or the RBD and the rest of the tested Hevein-like lectins (i.e., Smo443112, Smo99416, Smo437354, and Smo139127), hence, they were excluded from further analysis.
3.6. Hot-spot analysis of RBD-lectin complex
Using Knowledge-based FADE and Contacts 2 (KFC-2) the hotspot residues and/or clusters were predicted at the interface between S protein and lectins. The values from the KFC-a model were used as their predictability outcomes that were superior to those of the KFC-b model because it is based on interface solvation, atomic density, and plasticity features as indicated earlier [
49]. Specifically, residues Gln25, Asp21 from Smo446851, Smo125663 residues Arg91 and Gln11 and the Smo99732 Asp25 were identified as hot-spot residues and reported higher KFC-2-a scores compared to other residues (Supplementary File 1,
Figure S5). Complimentary hot-spot residues Val367, Asp364, Lys386, Gln115, and Thr109 were identified as hot-spot residues on S glycoprotein.
3.7. Normal-State Analyses via Torsional Coordinate-Association
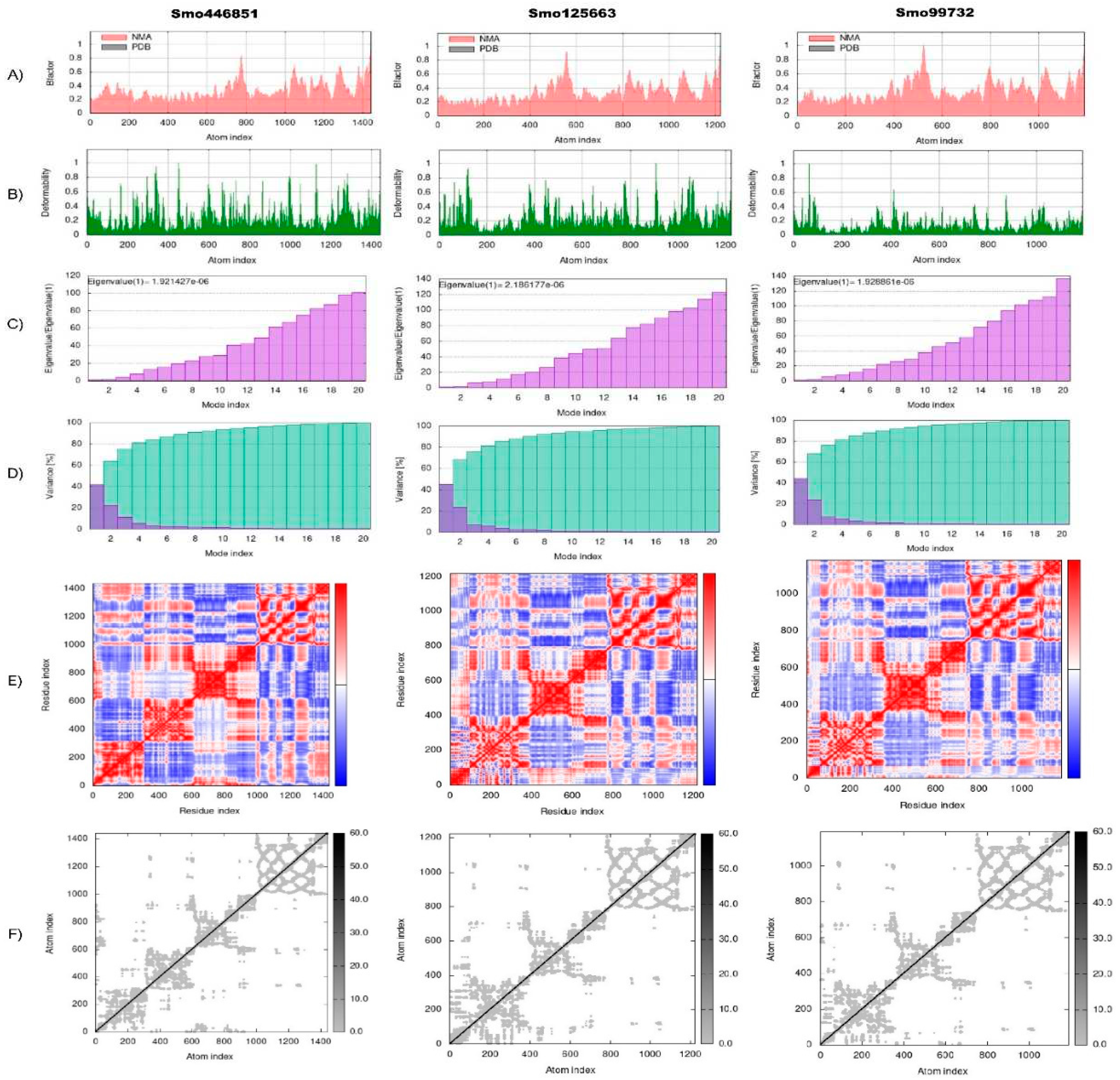
The torsion angle-related normal state analysis of lectins (Smo446851, Smo125663, and Smo99732) docked onto the Spike glycoprotein was performed using the iMODS server to examine their inherited stability and conformational mobility (
Figure 4). The B-factor is correlated to the atom’s displacement around conformational equilibrium. The three lectin complexes each with S-protein have the highest flexibility and large atomic displacements around the atomic positional range (400 – 600) (
Figure 4A). The complex deformability index which indicates higher individual distortions for the docked complexes showed general steady binding characterized by minimum deformities at coordination within the range (0 – 1 Å) and the peaks represent the location of hinges, Smo99732-S protein complex is more rigid than the other two complexes (
Figure 4B). Considering that the energy required to distort the complex is proportional to its eigenvalue, the lower the eigenvalue, the easier and the complex is to deform [
66]. The estimated eigenvalues representing the motion stiffness for each lectin-spike protein complex were 1.92 x 10-6, 2.12 x 10-6, and 1.93 x 10-6 for Smo446851, Smo125663, and Smo99732 complexes, respectively (
Figure 4C). The eigenvalues values were inversely proportional to variance predicting the significantly higher mobility of the lectins as compared to the spike protein across collective functional motion (
Figure 4D). The iMODS server provided a covariance matrix depicting uncorrelated (white), correlated (red), and anti-correlated (blue) residue pairs. The three docked complexes have high correlated residue-pair motion compared to the uncorrelated residue-pair motion (
Figure 4E). The elastic-network model describes the lectin-spike protein complex’s distinct flexibility patterns. It visualizes the atom pairs connected via springs by illustrating them according to their stiffness degrees. Dark grey is usually associated with stiffer strings. The dots indicate one spring, and a grey area indicates stiffer springs (
Figure 4F). Overall, the docked complexes showed stable binding, minimum deformities, and complex rigidity.
3.8. Mutant spike protein interaction with lectins
The carbohydrate-interacting lectins Smo446851, Smo125663, and Smo99732 were studied for interaction with different spike protein mutants commonly occur in SARS-CoV-2 variants (alpha, beta, gamma, delta and omicron) (
Figure 5). Five mutations related to the RBM region were selected and 5 mutant 6VXX structures were produced, one for each mutation. Analyzing the 15 generated docked complexes revealed that almost all the RBM regions were now accessible for the bind of Hevein-like lectins. And my forms of interactions including the formation of hydrogen bonds were evident. Ile468 which is one of the residues required for the binding of the ACE-2 receptor, this residue is mainly targeted by lectins in 11 complexes related to all mutations (Supplementary file 1
Table S4). However, Smo125663 and Smo99732 did not interact with N-glycan complexes and the interaction were solely in a form of protein-protein manner. Smo446851 retained its carbohydrate-protein interaction with NAG1307 in mutant Leu452Arg, and NAG1306 in mutants Thr478Lys and Asn501Lys (Supplementary file 1
Figure S6). The calculated MM/GBSA free binding energy for Smo446851-mutant Glu484Lys complex were higher than what is reported for parent RBD and other mutant complexes (-29.16 Kcal/mol). Smo125663 showed relatively poor interaction among the mutant complexes and compared to its interaction with parent RBD, while Smo99732 maintained the highest free binding energy score in all mutant complexes except for the Glu484Lys-RBD complex (-8.93 Kcal/mol).
Furthermore, the analysis of spike proteins that carries different sets of mutation points in their RBM that correspond to each variant (alpha (E501Y), beta (K417N, E484K, and E501Y) gamma (K417T, E484K, and E501Y), delta (L452R, T478K, and E484Q), and omicron (K417N, T478K, E484A, and E501Y)) (
Figure 5) revealed that Smo125663 have better free binding energy with both variants alpha (-31.8 Kcal/mol) and beta (-21.49 Kcal/mol) compared to the other lectins. It only interacts in a protein-protein manner with the RBD by forming six hydrogen bonds. The three lectins bind the RBD of the variant omicron in relatively low free binding energies. The carbohydrate-protein binding was observed for the interaction between Smo46851 active residue Thr49 of the lectin domain and the alpha mutatant S protein’s NAG1306 at 2.94 Å instead of the NAG1307 interaction reported for the parent spike protein (
Table 3).
4. Discussion
The spike moss Selaginella moellendorffii is a member of the lycophytes, which is classified under one of the three surviving families nowadays (Selaginellaceae). Its genome is the smallest reported plant genome with a size of ~100Mbp [
21]. The S. moellendorffii is a genetic model being utilized as a key to understanding how vascular plants evolved and gained many beneficial traits. Evolutionary studies showed that S. moellendorffii genome harbors genes members belonging to all documented plant lectin families except for the Agaricus bisporus agglutinin (ABA) family [
67]. The functional plasticity of plant lectins allows them to participate in various endogenous biological processes as well as defense molecules against predators and pathogens. Hence, their wide application in medicine and pharmaceutical fields. With the emergence of high throughput genome sequencing, the availability of genomic sources databases, datasets and their translation, and tools, novel sources of bioactive peptides and proteins were easy to identify and study. Hevein-like lectins are defense molecules, their activity is mediated by the interaction with pathogenic microorganisms’ surface chitin or its derivatives N-acetylglucosamine [
68]. Since the start of the COVID-19 pandemic, scientists have been searching for new drugs and vaccines to combat the virus. Many plant lectins inhibit SARS-CoV-2 by targeting the spike protein either through the interaction with the complex or high-mannose N-glycans found in the vicinity of the RBD or by interacting with the RBD residues through direct protein-protein binding [
16,
18]. Lectins from the Hevein (chitin-binding lectins) family mined form S. moellendorffii genome were studied for both binding modes to the RBD of the spike protein.
Three Hevein-like lectins (i.e., Smo35272, Smo425957, and Smo403798) bind in the RBD region of the spike protein that spans from residues 319 - 541 in a protein-protein manner. However, none of these lectins formed any type of interaction with the residues that constitute the ACE 2 receptor binding motif (residues 438 – 508, Tyr505, Gly502, Gly496, Asn501, Thr500, Gln498, Tyr449, Gly446, Tyr489, Phe456, Lys417, Gln493, Leu455, and Tyr453). These lectins bound deeply within the groove located at the top of the spike protein while some of the residues interacted with the amino acids flanking the RBM, especially in the region 300 – 399. Unlike the interaction reported for the Urtica dioica Agglutinin (UDA), examined for binding RBD (6VXX), UDA bound14 residues (i.e., Tyr505, Gly502, Gly496, Asn501, Thr500, Gln498, Tyr449, Gly446, Tyr489, Phe456, Lys417, Gln493, Leu455, and Tyr453) which are also involved in the RBD-ACE 2 interaction (Sabzian-Molaei et al., 2022). Therefore, the binding of Smo35272, Smo425957, and Smo403798 needs further investigation to understand how these lectins can influence and interfere with the binding of ACE 2.
Porto et al., (2012) studied the binding of Smo99732 (GenBank ID: XP 002973523) devoid of the signal peptide sequence (23 amino acids) with N, N, N- triacetyleglucosamine (GlcNAc)3. Three of these amino acids Phe20, Tyr22, and Tyr29 were responsible for the binding stabilized with hydrogen bonds during the most of simulation time [
69]. These residues are conserved as do part of the Hevein domain active site. However, the interaction with the RBD NAG1321 and NAG1307 via hydrogen bonds involved a different set of amino acids (Smo99732 – Phe43, Smo125663 – Ser30, Smo446851 – Ala26 and Arg29). The Asn343, along with Asn282 and Asn331 act as shield that covers and protects the spike protein’s RBD against neutralizing antibodies. They are also involved in the binding of the ACE2-RBD. Mutations or blocking of these glycosides are reported to influence viral binding and significantly reduce its infectivity [
9]. Lokhande et al (2022) investigated the binding of several lectins to the RBS of the S protein. Only NPA Narcissus pseudonarcissus Agglutinin and UDA a Hevein-like lectin interacted with GlcNAc complex N-glycans. However, UDA a hololectin with 2 tandemly arrayed hevein domains shared 34.62% homology with Smo125663. Both lectins interact with the N-glycan through the formation of hydrogen bonds between the first domain and the complex at a distance of 2.24 and 2.5 Å, respectively. However, UDA interacts with NAG1322, while Smo125663 interacts with NAG1321 and both complexes also interact with RBD Asn370 [
64].
Beta, gamma, delta, and omicron are classified as variants of concern (VOC) due to their increased transmissibility, disease severity, immune response impact, and drug efficacy [
70]. Several mutations occur in the RBM of the spike protein’s RBD region endowing the virus with enhanced ACE 2 receptor binding and/or better antibody evasion [
71,
72]. By neutralizing the free virions, Triticum vulgaris agglutinin (WGA) has been shown to prevent infection of SARS-CoV-2 and its variants alpha and beta [
73]. In an in vitro experiment employing Vero E6 cell lines, Griffithsin (GRFT) was found to bind to the spike protein and thereby preventing SARS-CoV-2 and its variants delta and omicron from entering the cells [
74]. The NTL-125, another lectin, was investigated for its high affinity binding to the RBD of several variations. Such binding comes about through the α-helical tail of the protein which interacts with glycan moieties and increases the binding strength [
75]. In light of our findings, Smo99732 appears to be a promising candidate for carbohydrate-protein interactions with wide-type spike glycoproteins, given its complex stability, low free energy compared to other S. moellendorffii Hevein-like lectins, and capacity for improved protein-protein interactions with mutant variants’ RBMs.
5. Conclusion
This comprehensive in-silico investigation provided novel contribution among other published studies and together confirmed the effectiveness of plant mannose- and GlcNAc-specific lectins against RNA viruses including HIV, SARS-CoV, and SARS-CoV-2. The latter two being inhibited by lectin-viral Spike N-linked glycoprotein interactions. In the current study, by genome-wide search, we report on ten novel chitin-specific Hevein-like lectins from Selaginella moellendorffii, three of which, Smo446851, Smo125663, and Smo99732, were able to interact favorably with the receptor binding domain (RBD) of the spike protein. The binding specificity of these lectin homologs with spike-RBD can be further investigated using in vivo systems like the vero cell line. Additionally, given the current SARS-CoV-2 mutation frequency, we anticipate that S. moellendorffii lectin-based medications specially Smo99732 might be a candidate against newly emerging variants, such as alpha, beta, gamma, and Omicron that are responsible for the rapid spread of COVID-19. Lectins are easier and highly feasible candidates than molecular vaccines albeit toxicological analysis to address potential reactions is imperative.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary file 1, Table S1: trRosetta templates used for lectin homology modelling, Table S2: Predicted TM-score for each lectin model, and Table S3: The binding site residues of the lectins. Supplementary file 1, Figure S1: The Ramachandran plots for the Hevein-like lectin model structures Figure S2-A and Figure S2-B: The quality and Z-score of the Hevein-like lectin structural models (five models), Figure S3: Prediction of the secondary structure of Hevein-like lectins from Selaginella moellendorffii. Hh: α-helices, Tt: β-turns, Cc: Random Coil, Ec: Extended strand.
Author Contributions
Conceptualization, E.H.E.K, M.E.M.O., A.A. and K.B.S.; Methodology, data curation and visualization, A.I.D., M.A.I.,M.A., and M.E.M.O.; software, M.E.M.O, A.I.D., E.H.E.K. K.B.S, A.A.,K.F.A.,F.A.,M.A.; Validation, A.A., K.B.S., K.F.A., F.A., M.A., and E.H.E.K; formal analysis, X.X.; investigation, K.B.S, A.A.,E.H.E.K., A.I.D.,M.A.I..; resources, K.B.S, A.A.,E.H.E.K., A.I.D.,M.A.I. K.F.A., F.A., M.A.; Writing original draft M.E.M.O, A.I.D., E.H.E.K.; writing—review and editing, K.B.S, A.A.,K.F.A.,F.A.,M.A.; Supervision, E.H.E.K., A.A., and K.B.S.; Project administration, A.A. and K.B.S.; Funding acquisition K.B.S., A.A., K.F.A., F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research project was funded by Scientific Research Deanship at the University of Ha’il- Saudi Arabia, through project number RG-21074.
Ethical approval and Institutional Review Board Statement
No specific permits were required because all repository data servers were publicly accessible. However, this study has been reviewed and Approval by the Research Ethical Committee (REC) of the University of Ha’il, dated 22/11/2021 under numbers H-2021-215, File H-2020-632-16160.
Data Availability Statement
All data generated and analyzed during this study are included in the main article and its supplementary data provided in the supplementary section with this manuscript. Genomic data of Selaginella moellendorffii, were publicly available and open freely accessible and all related UTR links were provided within the article under relevant mentions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization Dashboard. WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. https://covid19.who.int/ (accessed 2023-04-15).
- Said, K. B.; Alsolami, A.; Fathuldeen, A.; Alshammari, F.; Alhiraabi, W.; Alaamer, S.; Alrmaly, H.; Aldamadi, F.; Aldakheel, D. F.; Moussa, S.; Al Jadani, A.; Bashir, A. In-Silico Pangenomics of SARS-CoV-2 Isolates Reveal Evidence for Subtle Adaptive Expression Strategies, Continued Clonal Evolution, and Sub-Clonal Emergences, Despite Genome Stability. Microbiology Research 2021, Vol. 12, Pages 204-233 2021, 12(1), 204–233. [Google Scholar] [CrossRef]
- Jungreis, I.; Sealfon, R.; Kellis, M. SARS-CoV-2 Gene Content and COVID-19 Mutation Impact by Comparing 44 Sarbecovirus Genomes. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, N.; Alonazi, M. A.; Algarni, H. A.; Issrani, R.; Alanazi, S. H.; Alruwaili, M. K.; Alanazi, G. R.; Iqbal, A.; Khattak, O. Knowledge, Attitude and Practice towards the COVID-19 Pandemic: A Cross-Sectional Survey Study among the General Public in the Kingdom of Saudi Arabia. Vaccines (Basel) 2022, 10(11). [Google Scholar] [CrossRef] [PubMed]
- Redin, C.; Thorball, C. W.; Fellay, J. Host Genomics of SARS-CoV-2 Infection. Eur J Hum Genet 2022, 30(8), 908–914. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A. K.; Head, J. R.; Gould, C. F.; Carlton, E. J.; Remais, J. V. Environmental Factors Influencing COVID-19 Incidence and Severity. Annu Rev Public Health 2022, 43, 271–291. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 Spike Protein. Curr Opin Virol 2021, 50, 173–182. [Google Scholar] [CrossRef]
- Huang, H. C.; Lai, Y. J.; Liao, C. C.; Yang, W. F.; Huang, K. Bin; Lee, I. J.; Chou, W. C.; Wang, S. H.; Wang, L. H.; Hsu, J. M.; Sun, C. P.; Kuo, C. T.; Wang, J.; Hsiao, T. C.; Yang, P. J.; Lee, T. A.; Huang, W.; Li, F. A.; Shen, C. Y.; Lin, Y. L.; Tao, M. H.; Li, C. W. Targeting Conserved N-Glycosylation Blocks SARS-CoV-2 Variant Infection in Vitro. EBioMedicine 2021, 74. [Google Scholar] [CrossRef]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The Glycosylation in SARS-CoV-2 and Its Receptor ACE2. Signal Transduct Target Ther 2021, 6(1). [Google Scholar] [CrossRef]
- Li, C.; Zhou, H.; Guo, L.; Xie, D.; He, H.; Zhang, H.; Liu, Y.; Peng, L.; Zheng, L.; Lu, W.; Mei, Y.; Liu, Z.; Huang, J.; Wang, M.; Shu, D.; Ding, L.; Lang, Y.; Luo, F.; Wang, J.; Huang, B.; Huang, P.; Gao, S.; Chen, J.; Qian, C. N. Potential Inhibitors for Blocking the Interaction of the Coronavirus SARS-CoV-2 Spike Protein and Its Host Cell Receptor ACE2. J Transl Med 2022, 20(1). [Google Scholar] [CrossRef]
- Wang, G.; Yang, M. L.; Duan, Z. L.; Liu, F. L.; Jin, L.; Long, C. B.; Zhang, M.; Tang, X. P.; Xu, L.; Li, Y. C.; Kamau, P. M.; Yang, L.; Liu, H. Q.; Xu, J. W.; Chen, J. K.; Zheng, Y. T.; Peng, X. Z.; Lai, R. Dalbavancin Binds ACE2 to Block Its Interaction with SARS-CoV-2 Spike Protein and Is Effective in Inhibiting SARS-CoV-2 Infection in Animal Models. Cell Res 2021, 31(1), 17–24. [Google Scholar] [CrossRef]
- Peumans, W. J.; Van Damme, E. J. M. Lectins as Plant Defense Proteins. Plant Physiol 1995, 109(2), 347–352. [Google Scholar] [CrossRef]
- Konozy, E. H. E.; Osman, M. E. fadil M. Plant Lectin: A Promising Future Anti-Tumor Drug. Biochimie 2022, 202, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Konozy, E.; Osman, M.; Dirar, A. Plant Lectins as Potent Anti-Coronaviruses, Anti-Inflammatory, Antinociceptive and Antiulcer Agents. Saudi J Biol Sci 2022, 29(6). [Google Scholar] [CrossRef] [PubMed]
- Konozy, E. H. E.; Osman, M. E. fadil M.; Ghartey-Kwansah, G.; Abushama, H. M. The Striking Mimics between COVID-19 and Malaria: A Review. Front Immunol 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Lempp, F. A.; Soriaga, L. B.; Montiel-Ruiz, M.; Benigni, F.; Noack, J.; Park, Y. J.; Bianchi, S.; Walls, A. C.; Bowen, J. E.; Zhou, J.; Kaiser, H.; Joshi, A.; Agostini, M.; Meury, M.; Dellota, E.; Jaconi, S.; Cameroni, E.; Martinez-Picado, J.; Vergara-Alert, J.; Izquierdo-Useros, N.; Virgin, H. W.; Lanzavecchia, A.; Veesler, D.; Purcell, L. A.; Telenti, A.; Corti, D. Lectins Enhance SARS-CoV-2 Infection and Influence Neutralizing Antibodies. Nature 2021, 598(7880), 342–347. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Amaral, D.; Markovitz, D.; Pinto, L. The Use of Lectins as Tools to Combat SARS-CoV-2. Curr Pharm Des 2021, 27(41), 4212–4222. [Google Scholar] [CrossRef]
- Barre, A.; Van Damme, E. J. M.; Simplicien, M.; Le Poder, S.; Klonjkowski, B.; Benoist, H.; Peyrade, D.; Rougé, P. Man-Specific Lectins from Plants, Fungi, Algae and Cyanobacteria, as Potential Blockers for SARS-CoV, MERS-CoV and SARS-CoV-2 (COVID-19) Coronaviruses: Biomedical Perspectives. Cells 2021, 10(7). [Google Scholar] [CrossRef]
- Sabzian-Molaei, F.; Khalili, M. A. N.; Sabzian-Molaei, M.; Shahsavarani, H.; Pour, A. F.; Rad, A. M.; Hadi, A. Urtica Dioica Agglutinin: A Plant Protein Candidate for Inhibition of SARS-COV-2 Receptor-Binding Domain for Control of Covid19 Infection. PLoS One 2022, 17(7). [Google Scholar] [CrossRef]
- Setyawan, A. D. Setyawan, A. D. (PDF) Traditionally utilization of Selaginella; field research and literature review. https://www.researchgate.net/publication/267861685_Traditionally_utilization_of_Selaginella_field_research_and_literature_review (accessed 2023-04-15).
- Banks, J. A.; Nishiyama, T.; Hasebe, M.; Bowman, J. L.; Gribskov, M.; DePamphilis, C.; Albert, V. A.; Aono, N.; Aoyama, T.; Ambrose, B. A.; Ashton, N. W.; Axtell, M. J.; Barker, E.; Barker, M. S.; Bennetzen, J. L.; Bonawitz, N. D.; Chapple, C.; Cheng, C.; Correa, L. G. G.; Dacre, M.; DeBarry, J.; Dreyer, I.; Elias, M.; Engstrom, E. M.; Estelle, M.; Feng, L.; Finet, C.; Floyd, S. K.; Frommer, W. B.; Fujita, T.; Gramzow, L.; Gutensohn, M.; Harholt, J.; Hattori, M.; Heyl, A.; Hirai, T.; Hiwatashi, Y.; Ishikawa, M.; Iwata, M.; Karol, K. G.; Koehler, B.; Kolukisaoglu, U.; Kubo, M.; Kurata, T.; Lalonde, S.; Li, K.; Li, Y.; Litt, A.; Lyons, E.; Manning, G.; Maruyama, T.; Michael, T. P.; Mikami, K.; Miyazaki, S.; Morinaga, S. I.; TakashiMurata; Mueller-Roeber, B.; Nelson, D. R.; Obara, M.; Oguri, Y.; Olmstead, R. G.; Onodera, N.; Petersen, B. L.; Pils, B.; Prigge, M.; Rensing, S. A.; Riaño-Pachón, D. M.; Roberts, A. W.; Sato, Y.; Scheller, H. V.; Schulz, B.; Schulz, C.; Shakirov, E. V.; Shibagaki, N.; Shinohara, N.; Shippen, D. E.; Sørensen, I.; Sotooka, R.; Sugimoto, N.; Sugita, M.; Sumikawa, N.; Tanurdzic, M.; Theißen, G.; Ulvskov, P.; Wakazuki, S.; Weng, J. K.; Willats, W. W. G. T.; Wipf, D.; Wolf, P. G.; Yang, L.; Zimmer, A. D.; Zhu, Q.; Mitros, T.; Hellsten, U.; Loqué, D.; Otillar, R.; Salamov, A.; Schmutz, J.; Shapiro, H.; Lindquist, E.; Lucas, S.; Rokhsar, D.; Grigoriev, I. V. The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 2011, 332 (6032), 960–963. [CrossRef]
- Goodstein, D. M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R. D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; Rokhsar, D. S. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res 2012, 40. (Database issue). [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H. Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; Pesseat, S.; Quinn, A. F.; Sangrador-Vegas, A.; Scheremetjew, M.; Yong, S. Y.; Lopez, R.; Hunter, S. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30(9), 1236–1240. [Google Scholar] [CrossRef]
- Guo, A.-Y. ,; Zhu, Q.-H. ,; Chen, Z. ,; Luo, J.-C. [GSDS: a gene structure display server] - PubMed. https://pubmed.ncbi.nlm.nih.gov/17681935/ (accessed 2023-04-15).
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A. H. Gene Duplication and Evolution in Recurring Polyploidization-Diploidization Cycles in Plants. Genome Biol 2019, 20(1). [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H. R.; Frank, M. H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13(8), 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35(6), 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.; Efroni, I.; Rahni, R.; Birnbaum, K. D. The Selaginella Rhizophore Has a Unique Transcriptional Identity Compared with Root and Shoot Meristems. New Phytol 2018, 222(2), 882–894. [Google Scholar] [CrossRef] [PubMed]
- Ge, S. X.; Son, E. W.; Yao, R. IDEP: An Integrated Web Application for Differential Expression and Pathway Analysis of RNA-Seq Data. BMC Bioinformatics 2018, 19(1). [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P. X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res 2018, 46(W1), W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J. J.; Tsirigos, K. D.; Sønderby, C. K.; Petersen, T. N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat Biotechnol 2019, 37(4), 420–423. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E. L. L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J Mol Biol 2001, 305(3), 567–580. [Google Scholar] [CrossRef]
- Gupta, R. .; Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function - PubMed. https://pubmed.ncbi.nlm.nih.gov/11928486/ (accessed 2023-04-14).
- Steentoft, C.; Vakhrushev, S. Y.; Joshi, H. J.; Kong, Y.; Vester-Christensen, M. B.; Schjoldager, K. T. B. G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N. B.; Marcos-Silva, L.; Gupta, R.; Paul Bennett, E.; Mandel, U.; Brunak, S.; Wandall, H. H.; Levery, S. B.; Clausen, H. Precision Mapping of the Human O-GalNAc Glycoproteome through SimpleCell Technology. EMBO J 2013, 32(10), 1478–1488. [Google Scholar] [CrossRef]
- Horton, P.; Park, K. J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C. J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res 2007, 35. (Web Server issue). [Google Scholar] [CrossRef]
- Zhao, L.; Poschmann, G.; Waldera-Lupa, D.; Rafiee, N.; Kollmann, M.; Stühler, K. OutCyte: A Novel Tool for Predicting Unconventional Protein Secretion. Sci Rep 2019, 9(1). [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Comput Appl Biosci 1995, 11(6), 681–684. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The TrRosetta Server for Fast and Accurate Protein Structure Prediction. Nat Protoc 2021, 16(12), 5634–5651. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S. C.; Davis, I. W.; Arendall, W. B.; De Bakker, P. I. W.; Word, J. M.; Prisant, M. G.; Richardson, J. S.; Richardson, D. C. Structure Validation by Calpha Geometry: Phi,Psi and Cbeta Deviation. Proteins 2003, 50(3), 437–450. [Google Scholar] [CrossRef]
- Laskowski, R. A.; Jabłońska, J.; Pravda, L.; Vařeková, R. S.; Thornton, J. M. PDBsum: Structural Summaries of PDB Entries. Protein Sci 2018, 27(1), 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M. J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res 2007, 35. (Web Server issue). [Google Scholar] [CrossRef] [PubMed]
- Berman, H. M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T. N.; Weissig, H.; Shindyalov, I. N.; Bourne, P. E. The Protein Data Bank. Nucleic Acids Res 2000, 28(1), 235–242. [Google Scholar] [CrossRef] [PubMed]
- Walls, A. C.; Park, Y. J.; Tortorici, M. A.; Wall, A.; McGuire, A. T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181(2), 281–292e6. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G. A.; Dahlgren, M. K.; Russell, E.; Von Bargen, C. D.; Abel, R.; Friesner, R. A.; Harder, E. D. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J Chem Theory Comput 2021, 17(7), 4291–4300. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C. J.; Lu, S.; Chitsaz, F.; Derbyshire, M. K.; Geer, R. C.; Gonzales, N. R.; Gwadz, M.; Hurwitz, D. I.; Lu, F.; Marchler, G. H.; Song, J. S.; Thanki, N.; Wang, Z.; Yamashita, R. A.; Zhang, D.; Zheng, C.; Geer, L. Y.; Bryant, S. H. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res 2017, 45(D1), D200–D203. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S. Y. HDOCK: A Web Server for Protein-Protein and Protein-DNA/RNA Docking Based on a Hybrid Strategy. Nucleic Acids Res 2017, 45(W1), W365–W373. [Google Scholar] [CrossRef]
- Weng, G.; Wang, E.; Wang, Z.; Liu, H.; Zhu, F.; Li, D.; Hou, T. HawkDock: A Web Server to Predict and Analyze the Protein-Protein Complex Based on Computational Docking and MM/GBSA. Nucleic Acids Res 2019, 47(W1), W322–W330. [Google Scholar] [CrossRef] [PubMed]
- Biovia; Discovery Studio. BIOVIA Discovery Studio - BIOVIA - Dassault Systèmes®. https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (accessed 2023-04-15).
- Zhu, X.; Mitchell, J. C. KFC2: A Knowledge-Based Hot Spot Prediction Method Based on Interface Solvation, Atomic Density, and Plasticity Features. Proteins 2011, 79(9), 2671–2683. [Google Scholar] [CrossRef]
- López-Blanco, J. R.; Aliaga, J. I.; Quintana-Ortí, E. S.; Chacón, P. IMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res 2014, 42. (Web Server issue). [Google Scholar] [CrossRef] [PubMed]
- Lopéz-Blanco, J. R.; Garzón, J. I.; Chacón, P. IMod: Multipurpose Normal Mode Analysis in Internal Coordinates. Bioinformatics 2011, 27(20), 2843–2850. [Google Scholar] [CrossRef]
- Kezuka, Y.; Kojima, M.; Mizuno, R.; Suzuki, K.; Watanabe, T.; Nonaka, T. Structure of Full-Length Class I Chitinase from Rice Revealed by X-Ray Crystallography and Small-Angle X-Ray Scattering. Proteins 2010, 78(10), 2295–2305. [Google Scholar] [CrossRef]
- Morohashi, K.; Sasaki, K.; Sakabe, N. ,; Sakabe, K. RCSB PDB - 3W3E: Structure of Vigna unguiculata chitinase with regulation activity of the plant cell wall. https://www.rcsb.org/structure/3w3e (accessed 2023-04-15).
- Landim, P. G. C.; Correia, T. O.; Silva, F. D. A.; Nepomuceno, D. R.; Costa, H. P. S.; Pereira, H. M.; Lobo, M. D. P.; Moreno, F. B. M. B.; Brandão-Neto, J.; Medeiros, S. C.; Vasconcelos, I. M.; Oliveira, J. T. A.; Sousa, B. L.; Barroso-Neto, I. L.; Freire, V. N.; Carvalho, C. P. S.; Monteiro-Moreira, A. C. O.; Grangeiro, T. B. Production in Pichia Pastoris, Antifungal Activity and Crystal Structure of a Class I Chitinase from Cowpea (Vigna Unguiculata): Insights into Sugar Binding Mode and Hydrolytic Action. Biochimie 2017, 135, 89–103. [Google Scholar] [CrossRef]
- Balu, K. E.; Ramya, K. S.; Radha, A.; Krishnasamy, G. Structure of Intact Chitinase with Hevein Domain from the Plant Simarouba Glauca, Known for Its Traditional Anti-Inflammatory Efficacy. Int J Biol Macromol 2020, 161, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Hennig, M.; Schlesier, B.; Hohne, W. Structure of Jack Bean Chitinase. Acta Crystallogr D Biol Crystallogr 2000, 56 Pt 9, 1096–1099. [Google Scholar] [CrossRef]
- Kerff, F.; Amoroso, A.; Herman, R.; Sauvage, E.; Petrella, S.; Filée, P.; Charlier, P.; Joris, B.; Tabuchi, A.; Nikolaidis, N.; Cosgrove, D. J. Crystal Structure and Activity of Bacillus Subtilis YoaJ (EXLX1), a Bacterial Expansin That Promotes Root Colonization. Proc Natl Acad Sci U S A 2008, 105(44), 16876–16881. [Google Scholar] [CrossRef]
- Yennawar, N. H.; Li, A. C.; Dudzinski, D. M.; Tabuchi, A.; Cosgrove, D. J. Crystal Structure and Activities of EXPB1 (Zea m 1), a Beta-Expansin and Group-1 Pollen Allergen from Maize. Proc Natl Acad Sci U S A 2006, 103(40), 14664–14671. [Google Scholar] [CrossRef]
- Pedorov A. A.; Ball, T. ,; Leistler, B. ,; Valenta, R. ,; Almo, S. C. ,; Burley, S. K. RCSB PDB - 1N10: Crystal Structure of Phl p 1, a Major Timothy Grass Pollen Allergen. https://www.rcsb.org/structure/1n10 (accessed 2023-04-14).
- Yennawar, N. H. ,; Yennawar, H. P. ,; Georgelis, N. ,; Cosgrove, D. J. 4js7 - Crystal structure of D78N mutant apo form of clavibacter michiganensis expansin - Summary - Protein Data Bank Japan. https://pdbj.org/mine/summary/4js7 (accessed 2023-04-15).
- Chakraborty, N.; Besra, A.; Basak, J. Molecular Cloning of an Amino Acid Permease Gene and Structural Characterization of the Protein in Common Bean (Phaseolus Vulgaris L.). Mol Biotechnol 2020, 62 (3), 210–217. [CrossRef]
- Kumar, S.; Tsai, C. J.; Nussinov, R. Factors Enhancing Protein Thermostability. Protein Eng 2000, 13(3), 179–191. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Supekar, N. T.; Gleinich, A. S.; Azadi, P. Deducing the N- and O-Glycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. Glycobiology 2020, 30(12), 981–988. [Google Scholar] [CrossRef] [PubMed]
- Lokhande, K. B.; Apte, G. R.; Shrivastava, A.; Singh, A.; Pal, J. K.; Venkateswara Swamy, K.; Gupta, R. K. Sensing the Interactions between Carbohydrate-Binding Agents and N-Linked Glycans of SARS-CoV-2 Spike Glycoprotein Using Molecular Docking and Simulation Studies. J Biomol Struct Dyn 2022, 40(9), 3880–3898. [Google Scholar] [CrossRef] [PubMed]
- Itakura, Y.; Nakamura-Tsuruta, S.; Kominami, J.; Tateno, H.; Hirabayashi, J. Sugar-Binding Profiles of Chitin-Binding Lectins from the Hevein Family: A Comprehensive Study. Int J Mol Sci 2017, 18(6). [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Rizwan, M.; Zeb, A.; Eldeen, M. A.; Hassan, S.; Ur Rehman, A.; A. Eid, R.; Samir A. Zaki, M.; M. Albadrani, G.; E. Altyar, A.; Nouh, N. A. T.; Abdel-Daim, M. M.; Ullah, A. Identification of a Potential Vaccine against Treponema Pallidum Using Subtractive Proteomics and Reverse-Vaccinology Approaches. Vaccines (Basel) 2022, 11(1). [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E. J. M. Messages From the Past: New Insights in Plant Lectin Evolution. Front Plant Sci 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A. A.; Naumann, T. A.; Price, N. P. J.; Rogozhin, E. A.; Andreev, Y. A.; Vassilevski, A. A.; Odintsova, T. I. Novel Mode of Action of Plant Defense Peptides - Hevein-like Antimicrobial Peptides from Wheat Inhibit Fungal Metalloproteases. FEBS J 2014, 281(20), 4754–4764. [Google Scholar] [CrossRef]
- Porto, W. F.; Souza, V. A.; Nolasco, D. O.; Franco, O. L. In Silico Identification of Novel Hevein-like Peptide Precursors. Peptides (N.Y.) 2012, 38(1), 127–136. [Google Scholar] [CrossRef]
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 Variants of Concern and Potential Intervention Approaches. Crit Care 2021, 25(1). [Google Scholar] [CrossRef]
- Harvey, W. T.; Carabelli, A. M.; Jackson, B.; Gupta, R. K.; Thomson, E. C.; Harrison, E. M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S. J.; Robertson, D. L. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat Rev Microbiol 2021, 19(7), 409–424. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J. W.; Zhu, X.; Srivastava, S. S.; Berezuk, A. M.; Zhou, S.; Tuttle, K. S.; Kim, A.; Li, W.; Dimitrov, D. S.; Subramaniam, S. Structural Analysis of Receptor Binding Domain Mutations in SARS-CoV-2 Variants of Concern That Modulate ACE2 and Antibody Binding. Cell Rep 2021, 37(12). [Google Scholar] [CrossRef] [PubMed]
- Auth, J.; Fröba, M.; Große, M.; Rauch, P.; Ruetalo, N.; Schindler, M.; Morokutti-kurz, M.; Graf, P.; Dolischka, A.; Prieschl-grassauer, E.; Setz, C.; Schubert, U. Lectin from Triticum Vulgaris (WGA) Inhibits Infection with SARS-CoV-2 and Its Variants of Concern Alpha and Beta. Int J Mol Sci 2021, 22(19). [Google Scholar] [CrossRef] [PubMed]
- Ahan, R. E.; Hanifehnezhad, A.; Kehribar, E.; Oguzoglu, T. C.; Földes, K.; Özçelik, C. E.; Filazi, N.; Öztop, S.; Palaz, F.; Önder, S.; Bozkurt, E. U.; Ergünay, K.; Özkul, A.; Şeker, U. Ö. Ş. A Highly Potent SARS-CoV-2 Blocking Lectin Protein. ACS Infect Dis 2022, 8(7), 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Paul, S.; Singh, C.; Chowdhury, N.; Nag, P.; Das, S.; Kumar, S.; Sharma, A.; Das, D. K.; Dutta, D.; Thakur, K. G.; Bagchi, A.; Shriti, S.; Das, K. P.; Ringe, R. P.; Das, S. A Novel Plant Lectin, NTL-125, Interferes with SARS-CoV-2 Interaction with HACE2. Virus Res 2022, 315. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).