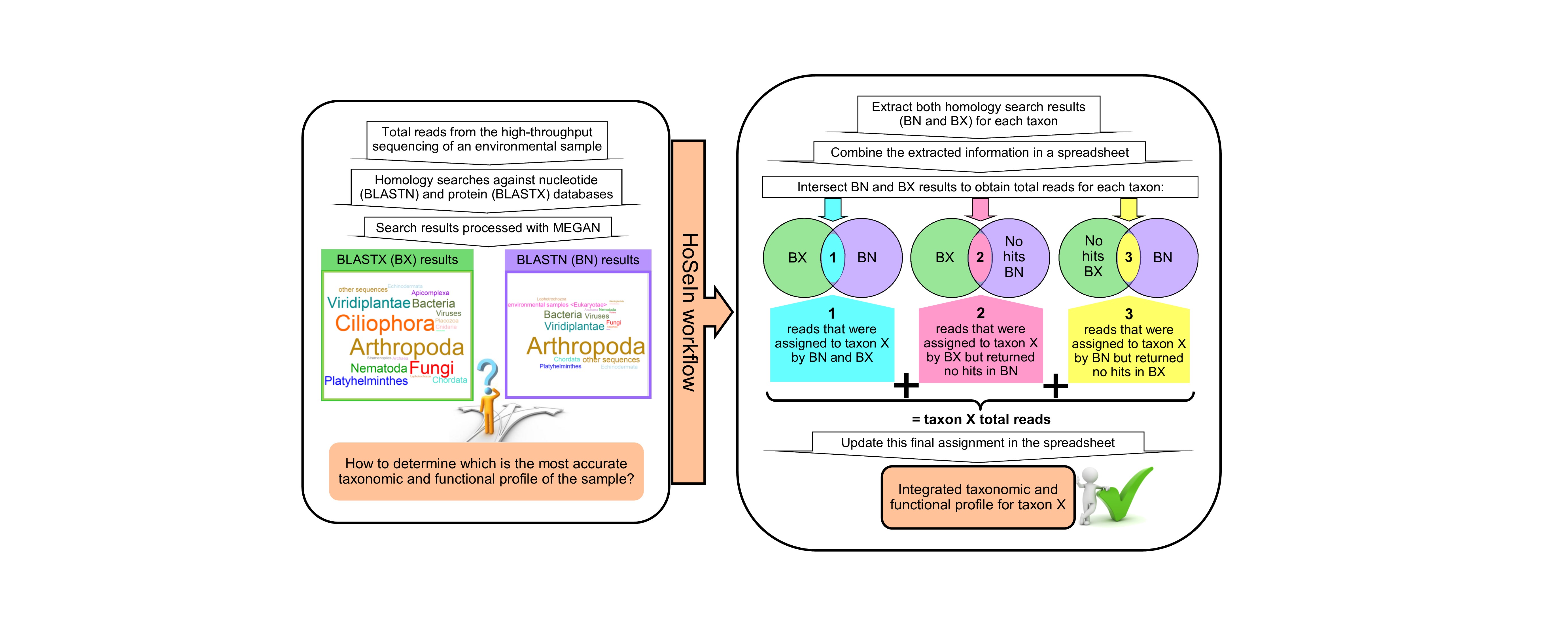
Data generated by metagenomic and metatranscriptomic experiments is both enormous and inherently noisy [1]. When using taxonomy-dependent alignment-based methods to classify and label reads, such as MEGAN [2], the first step consists in performing homology searches against sequence databases. To obtain the most information from the samples, nucleotide sequences are usually compared to various databases (i.e., nucleotide and protein) using local sequence aligners such as BLASTN and BLASTX [3]. Nevertheless, the analysis and integration of these results can be problematic because the outputs from these searches usually show differences, which can be notorious when working with RNA-seq (Personal observation; Graphical abstract). These inconsistencies led us to develop the HoSeIn workflow to determine the unequivocal taxonomic and functional profile of environmental samples, based on the assumption that the sequences that correspond to a certain taxon are composed of (Graphical abstract): 1) sequences that were assigned to the same taxon by both homology searches, plus 2) sequences that were assigned to that taxon by one of the homology searches but returned no hits in the other one, and vice versa.