Article
Version 1
Preserved in Portico This version is not peer-reviewed
Prokaryotic gene-family size correlations
Version 1
: Received: 29 July 2016 / Approved: 29 July 2016 / Online: 29 July 2016 (16:24:29 CEST)
A peer-reviewed article of this Preprint also exists.
Carmi, G.; Bolshoy, A. Gene-Family Extension Measures and Correlations. Life 2016, 6, 30. Carmi, G.; Bolshoy, A. Gene-Family Extension Measures and Correlations. Life 2016, 6, 30.
Abstract
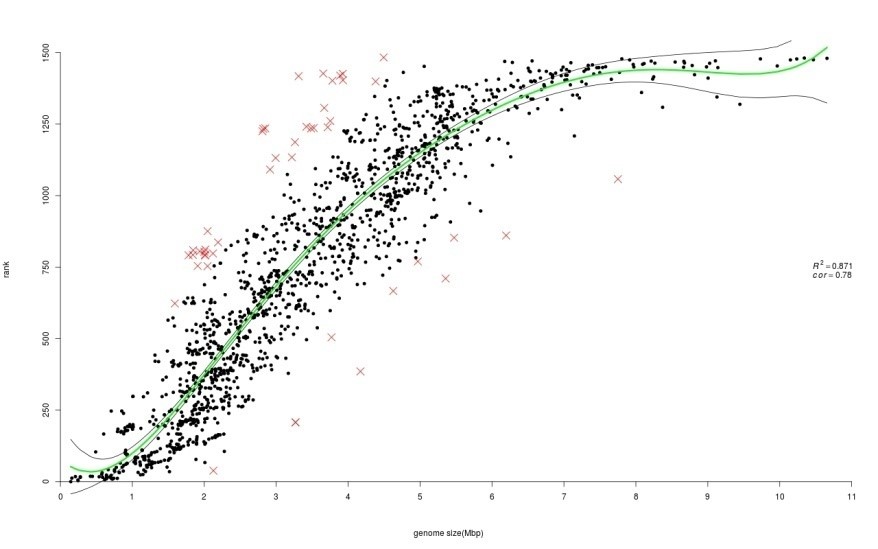
The existence of multiple copies of genes is a well-known phenomenon. A gene family is a set of sufficiently similar genes, formed by gene duplication. In earlier works conducted on limited number of completely sequenced and annotated genomes it was found that size of gene family and size of genome are positively correlated. Additionally, it was found that several atypical microbes deviated from the observed general trend. In this study, we reexamined these associations on a larger dataset consisting of 1484 prokaryotic genomes and using several ranking approaches. We applied ranking methods in such a way that genomes with lower number of paralogs would have lower rank. Until now only simple ranking methods were used; we applied the Kemeny optimal aggregation approach as well. Regression and correlation analysis were utilized in order to accurately quantify and characterize the relationships between measures of paralog indices and genome size. In addition, boxplot analysis was employed as a method for outlier detection. We found that, in general, all paralog indexes positively correlate with an increase of genome size. As expected, different groups of atypical prokaryotic genomes were found for different types of paralog quantities.
Keywords
number of paralogs; comparative genomics; combinatorial optimization; Mycoplasmas; Halophiles; Orientia; Mycobacterium leprae; genome size
Subject
Biology and Life Sciences, Biochemistry and Molecular Biology
Copyright: This is an open access article distributed under the Creative Commons Attribution License which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Comments (0)
We encourage comments and feedback from a broad range of readers. See criteria for comments and our Diversity statement.
Leave a public commentSend a private comment to the author(s)
* All users must log in before leaving a comment