Submitted:
21 January 2025
Posted:
22 January 2025
You are already at the latest version
Abstract
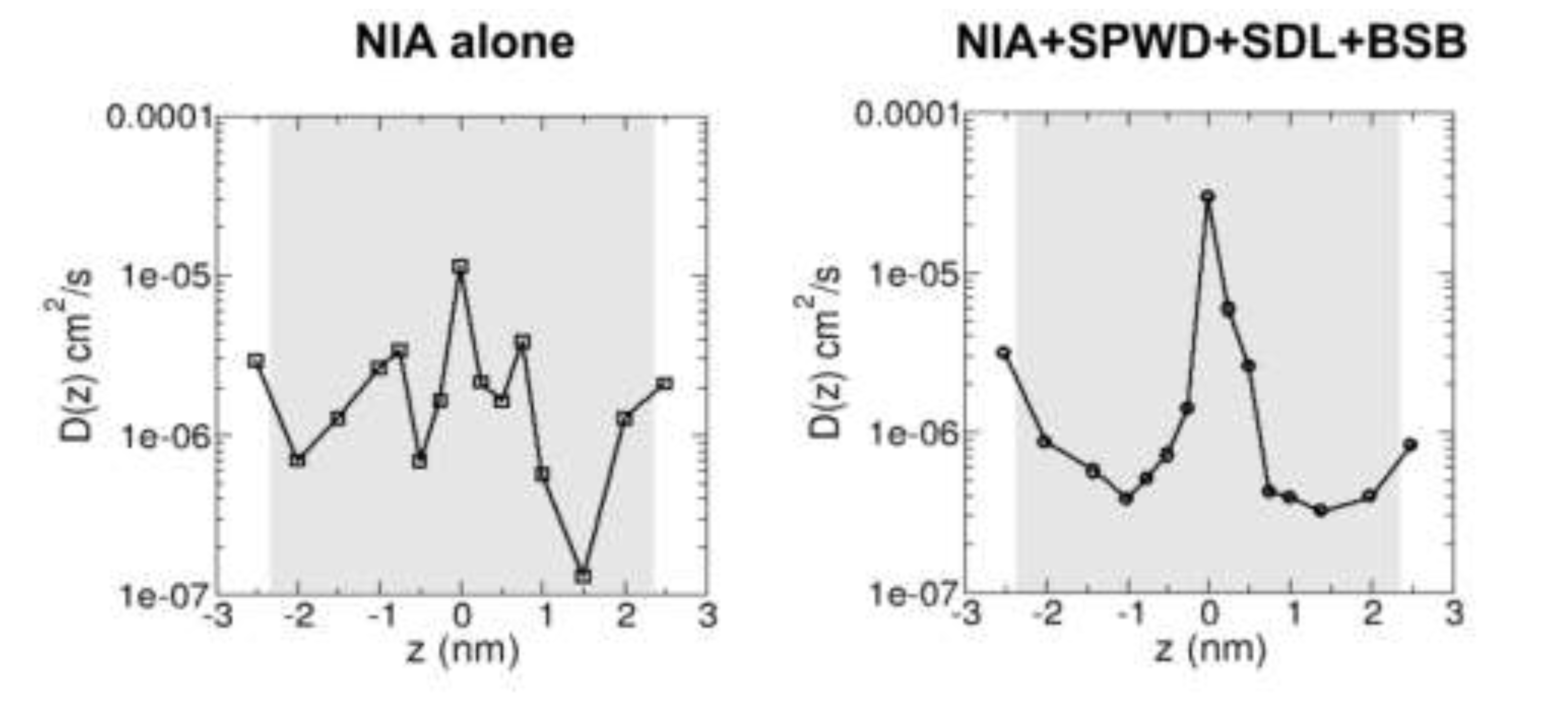
Niacinamide, a derivative of vitamin B3, has been shown to reduce skin pigmentation (i.e. acting as a brightening agent) and inflammatory responses such as dermatitis and acne vulgaris. However, niacinamide is a hydrophilic compound and poor partitioning to the lipid matrix in the uppermost layer of the skin (the stratum corneum or SC) limits its delivery to the skin. This necessitates the use of penetration enhancers to increase its bio-availability. In this study, we used computer simulations to investigate the skin penetration of niacinamide alone and in combination with other brightening agents that are also shown to be skin penetration enhancers, namely Sepiwhite®, bisabolol or sucrose dilaurate. Molecular dynamics simulations were performed to reveal molecular interactions of these brightening agents with a lipid bilayer model that mimics the SC lipid matrix. We observed minimal penetration of niacinamide into the SC lipid bilayer when applied alone or in combination with any one of the three compounds. However, when all three compounds were combined, a notable increase in penetration was observed. We showed 32% increase in the niacinamide diffusivity in the presence of other three brightening agents, which also work as penetration enhancer for niacinamide. These findings suggest that formulations containing multiple brightening agents, which works as penetration enhancers, may improve skin penetration of niacinamide and enhance the effectiveness of the treatment.
Keywords:
1. Introduction
2. Results
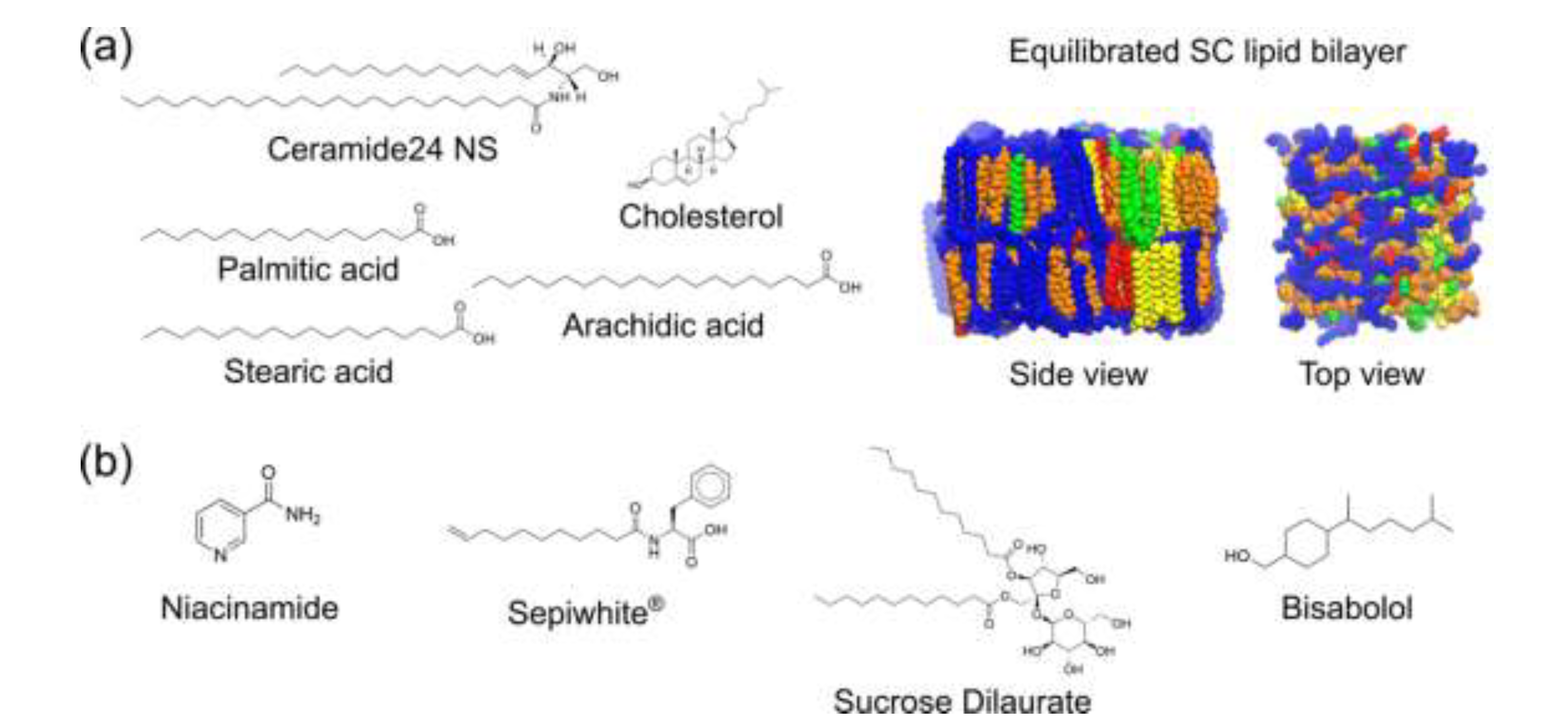
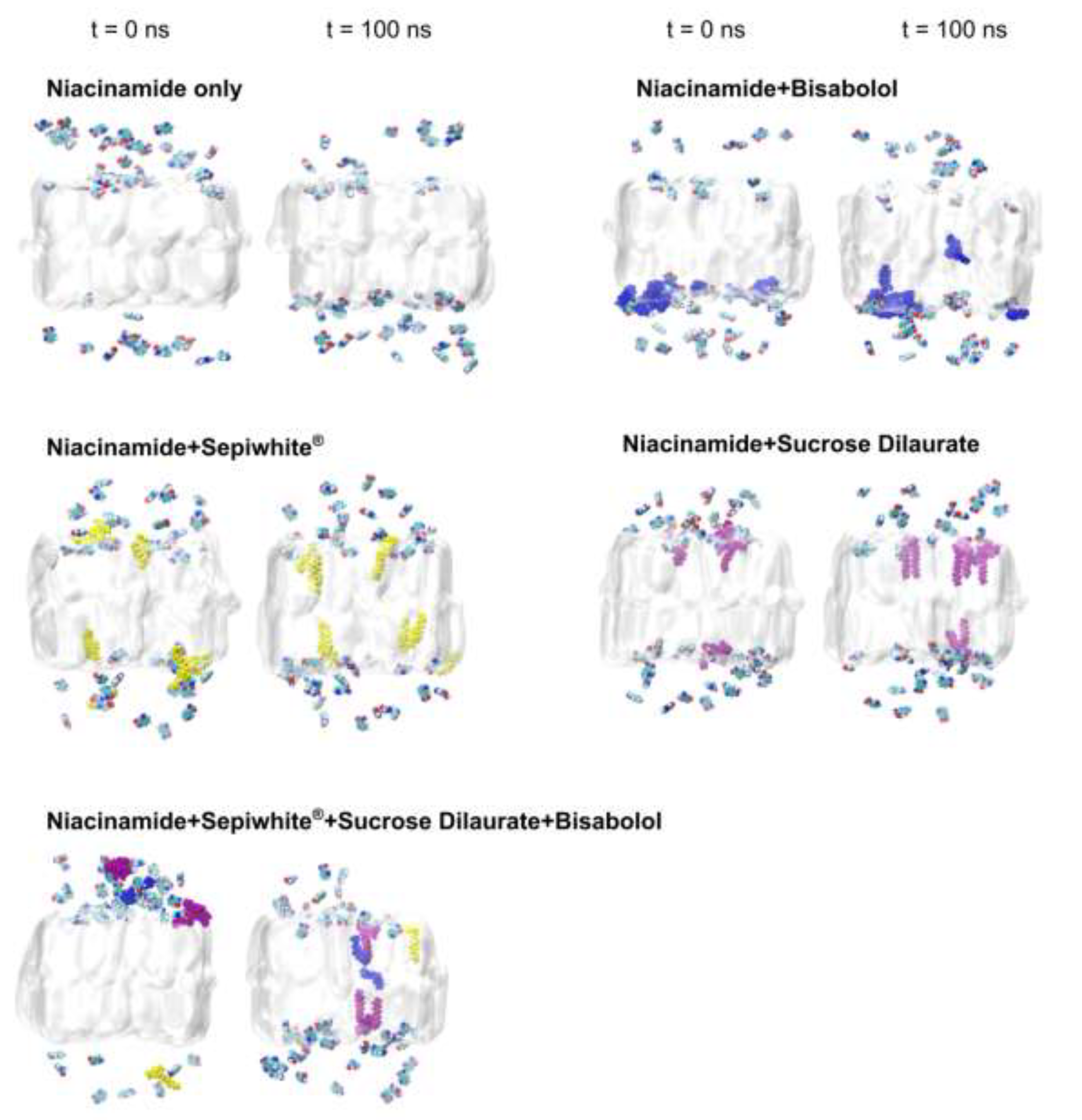
2.1. Brightening Agents Adsorb onto the SC Lipid Bilayer
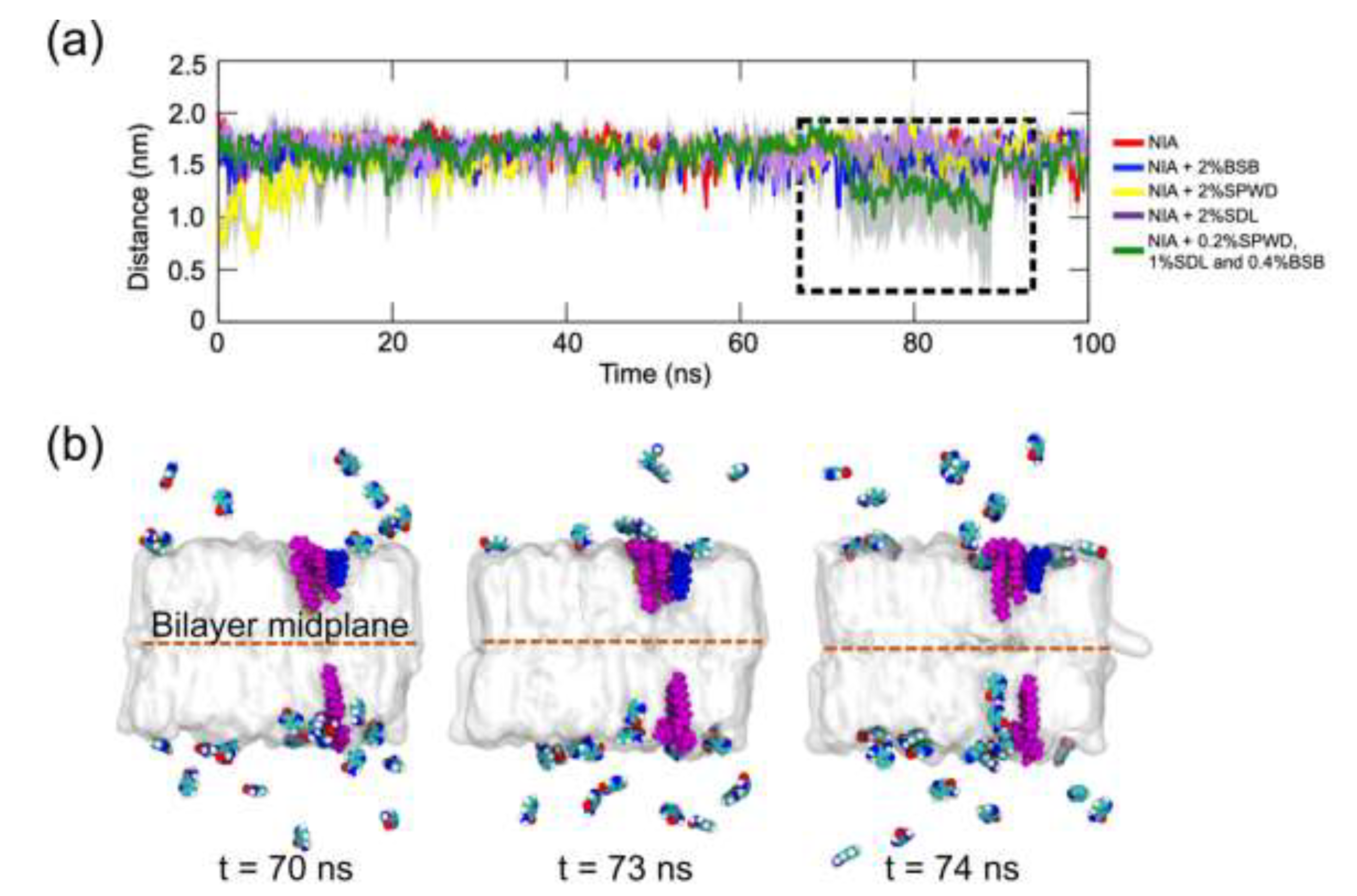
2.2. Brightening Agents Absorbed onto SC Lipid Bilayer Enhance Diffusivity of Niacinamide Across the Bilayer
3. Discussion
4. Materials and Methods
4.1. Generation and Equilibration of SC Lipid Bilayer Model
4.2. Calculation of Biophysical Properties of the SC Bilayer
4.3. Generation of brightening agent models and simulation of brightening agent-SC lipid bilayer interactions
4.4. Calculation of Position-Dependent Diffusivity of Niacinamide Across the SC Lipid Bilayer
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Matts, P.J.; Oblong, J.E.; Bissett, D.L. Int Fed Soc Cosmet Chem Mag. 2002, pp. 285–289.
- Boo, Y.C. Mechanistic Basis and Clinical Evidence for the Applications of Nicotinamide (Niacinamide) to Control Skin Aging and Pigmentation. Antioxidants 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Castanedo-Cazares, J.P.; Lárraga-Piñones, G.; Ehnis-Pérez, A.; Fuentes-Ahumada, C.; Oros-Ovalle, C.; Smoller, B.R.; Torres-Álvarez, B. Topical Niacinamide 4% and Desonide 0.05% for Treatment of Axillary Hyperpigmentation: A Randomized, Double-Blind, Placebo-Controlled Study. Clin Cosmet Investig Dermatol 2013, 6, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Bowes, J.; Piper, J.; Thiemermann, C. Inhibitors of the Activity of Poly (ADP-Ribose) Synthetase Reduce the Cell Death Caused by Hydrogen Peroxide in Human Cardiac Myoblasts. Br J Pharmacol 1998, 124, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Tanno, O.; Ota, Y.; Kitamura, N.; Inoue, S. Nicotinamide Increases Biosynthesis of Ceramides as Well as Other Stratum Corneum Lipids to Improve the Epidermal Permeability Barrier. British Journal of Dermatology 2000, 143, 524–531. [Google Scholar] [CrossRef]
- Khodaeiani, E.; Fouladi, R.F.; Amirnia, M.; Saeidi, M.; Karimi, E.R. Topical 4% Nicotinamide vs. 1% Clindamycin in Moderate Inflammatory Acne Vulgaris. Int J Dermatol 2013, 52, 999–1004. [Google Scholar] [CrossRef]
- Zhu, J.R.; Wang, J.; Wang, S.S. A Single-Center, Randomized, Controlled Study on the Efficacy of Niacinamide-Containing Body Emollients Combined with Cleansing Gel in the Treatment of Mild Atopic Dermatitis. Skin Research and Technology 2023, 29. [Google Scholar] [CrossRef]
- Hakozaki, T.; Minwalla, L.; Zhuang, J.; Chhoa, M.; Matsubara, A.; Miyamoto, K.; Greatens, A.; Hillebrand, G.G.; Bissett, D.L.; Boissy, R.E. The Effect of Niacinamide on Reducing Cutaneous Pigmentation and Suppression of Melanosome Transfer. British Journal of Dermatology 2002, 147, 20–31. [Google Scholar] [CrossRef]
- Greatens, A.; Hakozaki, T.; Koshoffer, A.; Epstein, H.; Schwemberger, S.; Babcock, G.; Bissett, D.; Takiwaki, H.; Arase, S.; Wickett, R.R.; et al. Effective Inhibition of Melanosome Transfer to Keratinocytes by Lectins and Niacinamide Is Reversible. Exp Dermatol 2005, 14, 498–508. [Google Scholar] [CrossRef]
- Khodaeiani, E.; Fouladi, R.F.; Amirnia, M.; Saeidi, M.; Karimi, E.R. Topical 4% Nicotinamide vs. 1% Clindamycin in Moderate Inflammatory Acne Vulgaris. Int J Dermatol 2013, 52, 999–1004. [Google Scholar] [CrossRef]
- Zhu, J.R.; Wang, J.; Wang, S.S. A Single-Center, Randomized, Controlled Study on the Efficacy of Niacinamide-Containing Body Emollients Combined with Cleansing Gel in the Treatment of Mild Atopic Dermatitis. Skin Research and Technology 2023, 29. [Google Scholar] [CrossRef]
- Fabbrocini, G.; Cantelli, M.; Monfrecola, G. Topical Nicotinamide for Seborrheic Dermatitis: An Open Randomized Study. Journal of Dermatological Treatment 2014, 25, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Ungerstedt, J.S.; Blombäck, M.; Söderström, T. 2003.
- Elias, P.M. Epidermal Lipids, Barrier Function, and Desquamation. Journal of Investigative Dermatology 1983, 80, S44–S49. [Google Scholar] [CrossRef]
- Elias, P.M.; Goerke, J.; Friend, D.S. Mammalian Epidermal Barrier Layer Lipids: Composition and Influence on Structure. Journal of Investigative Dermatology 1977, 69, 535–546. [Google Scholar] [CrossRef]
- Brown, M.B.; Martin, G.P.; Jones, S.A.; Akomeah, F.K. Dermal and Transdermal Drug Delivery Systems: Current and Future Prospects. Drug Delivery: Journal of Delivery and Targeting of Therapeutic Agents 2006, 13, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, D.; Matts, P.J.; Hadgraft, J.; Lane, M.E. In Vitro-in Vivo Correlation in Skin Permeation. Pharm Res 2014, 31, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kung, C.P.; Sil, B.C.; Lane, M.E.; Hadgraft, J.; Heinrich, M.; Sinko, B. Topical Delivery of Niacinamide: Influence of Binary and Ternary Solvent Systems. Pharmaceutics 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, T.; Nokhodchi, A.; Kaialy, W. Surfactants as Penetration Enhancers for Dermal and Transdermal Drug Delivery. In Percutaneous Penetration Enhancers Chemical Methods in Penetration Enhancement: Modification of the Stratum Corneum; Dragicevic, N., Maibach, H.I., Eds.; Springer Berlin Heidelberg, 2015; pp. 207–230 ISBN 9783662470398.
- Sohn, J.S.; Choi, J.S. Development and Evaluation of Niacinamide Transdermal Formulation by Artificial Membrane Permeability. Saudi Pharmaceutical Journal 2023, 31, 1229–1236. [Google Scholar] [CrossRef]
- Gupta, R.; Sridhar, D.B.; Rai, B. Molecular Dynamics Simulation Study of Permeation of Molecules through Skin Lipid Bilayer. Journal of Physical Chemistry B 2016, 120, 8987–8996. [Google Scholar] [CrossRef]
- Lundborg, M.; Wennberg, C.L.; Narangifard, A.; Lindahl, E.; Norlén, L. Predicting Drug Permeability through Skin Using Molecular Dynamics Simulation. Journal of Controlled Release 2018, 283, 269–279. [Google Scholar] [CrossRef]
- Wennberg, C.; Lundborg, M.; Lindahl, E.; Norlén, L. Understanding Drug Skin Permeation Enhancers Using Molecular Dynamics Simulations. J Chem Inf Model 2023, 63, 4900–4911. [Google Scholar] [CrossRef]
- Mistry, J.; Notman, R. Mechanisms of the Drug Penetration Enhancer Propylene Glycol Interacting with Skin Lipid Membranes. Journal of Physical Chemistry B 2024, 128, 3885–3897. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Card, P.D.; Chen, J.; Li, L.; Laughlin, T.; Jarrold, B.; Zhao, W.; Benham, A.M.; Määttä, A.T.; Hawkins, T.J.; et al. A Potential Role of Keratinocyte-Derived Bilirubin in Human Skin Yellowness and Its Amelioration by Sucrose Laurate/Dilaurate. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Csizmazia, E.; Erős, G.; Berkesi, O.; Berkó, S.; Szabó-Révész, P.; Csányi, E. Penetration Enhancer Effect of Sucrose Laurate and Transcutol on Ibuprofen. J. Drug Del. Sci. Tech. 2011, 21, 411–415. [Google Scholar] [CrossRef]
- Bolzinger, M.A.; Carduner, C.; Poelman, M.C. Bicontinuous Sucrose Ester Microemulsion: A New Vehicle for Topical Delivery of Niflumic Acid. Int J Pharm 1998, 176, 39–45. [Google Scholar] [CrossRef]
- Walters, K.A.; Florence, A.T.; Dugard, P.H. Interaction of Polyoxyethylene Alkyl Ethers with Cholesterol Monolayers 1. J Colloid Interface Sci 1982, 89. [Google Scholar] [CrossRef]
- Kabiri, H.; Tayarani-Najaran, Z.; Rahmanian-devin, P.; Vaziri, M.S.; Nasirizadeh, S.; Golmohammadzadeh, S.; Kamali, H. Preparation, Characterization, and Evaluation of Anti-Tyrosinase Activity of Solid Lipid Nanoparticles Containing Undecylenoyl Phenylalanine (Sepiwhite®). J Cosmet Dermatol 2022, 21, 6061–6071. [Google Scholar] [CrossRef]
- Katoulis, A.; Alevizou, A.; Soura, E.; Mantas, N.; Bozi, E.; Gregoriou, S.; Makris, M.; Rigopoulos, D. A Double-Blind Vehicle-Controlled Study of a Preparation Containing Undecylenoyl Phenylalanine 2% in the Treatment of Melasma in Females. J Cosmet Dermatol 2014, 13, 86–90. [Google Scholar] [CrossRef]
- Bissett, D.L.; Robinson, L.R.; Raleigh, P.S.; Miyamoto, K.; Hakozaki, T.; Li, J.; Kelm, G.R.; Johnson, M. Reduction in the Appearance of Facial Hyperpigmentation by Topical N-Undecyl-10-Enoyl-L-Phenylalanine and Its Combination with Niacinamide. J Cosmet Dermatol 2009, 8, 260–266. [Google Scholar] [CrossRef]
- Forrer, M.; Kulik, E.M.; Filippi, A.; Waltimo, T. The Antimicrobial Activity of Alpha-Bisabolol and Tea Tree Oil against Solobacterium Moorei, a Gram-Positive Bacterium Associated with Halitosis. Arch Oral Biol 2013, 58, 10–16. [Google Scholar] [CrossRef]
- De, O. Leite, G.; Leite, L.H.I.; De S. Sampaio, R.; Araruna, M.K.A.; De Menezes, I.R.A.; Da Costa, J.G.M.; Campos, A.R. (-)-α-Bisabolol Attenuates Visceral Nociception and Inflammation in Mice. Fitoterapia 2011, 82, 208–211. [Google Scholar] [CrossRef]
- Maurya, A.K.; Singh, M.; Dubey, V.; Srivastava, S.; Luqman, S.; Bawankule, D.U. α-(-)-Bisabolol Reduces Pro-Inflammatory Cytokine Production and Ameliorates Skin Inflammation. Curr Pharm Biotechnol 2014, 15, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jun, H.; Jung, E.; Ha, J.; Park, D. Whitening Effect of α-Bisabolol in Asian Women Subjects. Int J Cosmet Sci 2010, 32, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Kadir, R.; Barry, B.W. Alpha-Bisabolol, a Possible Safe Penetration Enhancer for Dermal and Transdermal Therapeutics. Int J Pharm 1991, 70, 87–94. [Google Scholar] [CrossRef]
- Chng, C.P.; Zhang, L.; Gupta, S.; Huang, C. Palmitoylation Enhances Short Polar Peptide Permeation across Stratum Corneum Lipid Bilayer: A Molecular Dynamics Study. Extreme Mech Lett 2024, 71, 102213. [Google Scholar] [CrossRef]
- Martinez, L.; Andrade, R.; Birgin, E.G.; Martinez, J. Packmol: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J Comput Chem 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J Comput Chem 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Páll, S.; Abraham, M.J.; Kutzner, C.; Hess, B.; Lindahl, E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In Proceedings of the Solving software challenges for exascale; Markidis, S., Laure, E., Eds.; Springer, Cham; 2015; pp. 3–27. [Google Scholar]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J Mol Graph 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Buchoux, S. FATSLiM: A Fast and Robust Software to Analyze MD Simulations of Membranes. Bioinformatics 2017, 33, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI Ligand Reader and Modeler for CHARMM Force Field Generation of Small Molecules. J Comput Chem 2017, 38, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J Comput Chem 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
| Brightening agent | ALogP | Molar volume (m3/mol) |
|---|---|---|
| Niacinamide | –0.32 | 322 |
| Sepiwhite® | 5.19 | 852 |
| Sucrose Dilaurate | 5.99 | 1607 |
| Bisabolol | 4.31 | 593 |
| Brightening agent(s) added | Bilayer thickness (nm) | Area/lipid (nm2) | # of lipid head-group H-bonds |
|---|---|---|---|
| None | 4.53 ± 0.02 | 0.99 ± 0.004 | 33.67 ± 0.21 |
| Niacinamide (NIA) | 4.55 ± 0.00 | 0.98 ± 0.007 | 35.84 ± 1.18 |
| NIA + 2% BSB | 4.54 ± 0.00 | 1.01 ± 0.000 | 37.34 ± 0.94 |
| NIA + 2% SPWD | 4.51 ± 0.01 | 1.01 ± 0.000 | 34.74 ± 1.32 |
| NIA + 2% SDL | 4.54 ± 0.01 | 0.99 ± 0.002 | 36.72 ± 0.40 |
| NIA + 0.2% SPWD, 1% SDL and 0.4% BSB | 4.51 ± 0.01 | 1.01 ± 0.021 | 37.48 ± 1.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





